+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human PSS2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PS lipid / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCDP-diacylglycerol-serine O-phosphatidyltransferase activity / L-serine-phosphatidylethanolamine phosphatidyltransferase / L-serine-phosphatidylethanolamine phosphatidyltransferase activity / Synthesis of PS / phosphatidylserine biosynthetic process / transferase activity / endoplasmic reticulum membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Li DY / Li XC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Molecular insights into human phosphatidylserine synthase 2 and its regulation of SREBP pathways. Authors: Dongyu Li / Hongwen Chen / Goncalo Vale / Nadia Elghobashi-Meinhardt / Alexandra Hatton / Shunxing Rong / Jeffrey G McDonald / Xiaochun Li /   Abstract: Homologous proteins share similar sequences, enabling them to work together in cells to support normal physiological functions. Phosphatidylserine synthases 1 and 2 (PSS1 and PSS2) are homologous ...Homologous proteins share similar sequences, enabling them to work together in cells to support normal physiological functions. Phosphatidylserine synthases 1 and 2 (PSS1 and PSS2) are homologous enzymes that catalyze the synthesis of phosphatidylserine (PS) from different substrates. PSS2 shows a preference for phosphatidylethanolamine (PE) as its substrate, whereas PSS1 can utilize either PE or phosphatidylcholine. Previous studies showed that inhibiting PSS1 promotes SREBP-2 cleavage. Interestingly, despite their homology, our findings reveal that PSS2 exerts an opposing effect on the cleavage of both SREBP-1 and SREBP-2. We resolved the cryo-electron microscopy (cryo-EM) structure of human PSS2 at 3.3 Å resolution. Structural comparison of the catalytic cavities between PSS1 and PSS2 along with molecular dynamics simulations uncovers the molecular details behind the substrate preference of PSS2 for PE. The lipidomic analysis showed that PSS2 deficiency leads to PE accumulation in the endoplasmic reticulum, which has been shown to inhibit the cleavage of sterol regulatory element-binding proteins (SREBPs) in mice. Thus, our findings reveal the intricate network of intracellular phospholipid metabolism and underscore the distinct regulatory roles of homologous proteins in cellular activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48797.map.gz emd_48797.map.gz | 167.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48797-v30.xml emd-48797-v30.xml emd-48797.xml emd-48797.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48797_fsc.xml emd_48797_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_48797.png emd_48797.png | 78.8 KB | ||
| Filedesc metadata |  emd-48797.cif.gz emd-48797.cif.gz | 6.3 KB | ||
| Others |  emd_48797_half_map_1.map.gz emd_48797_half_map_1.map.gz emd_48797_half_map_2.map.gz emd_48797_half_map_2.map.gz | 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48797 http://ftp.pdbj.org/pub/emdb/structures/EMD-48797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48797 | HTTPS FTP |
-Related structure data
| Related structure data |  9n0xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_48797.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48797.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.738 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_48797_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_48797_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of human PSS2
| Entire | Name: Structure of human PSS2 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of human PSS2
| Supramolecule | Name: Structure of human PSS2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylserine synthase 2
| Macromolecule | Name: Phosphatidylserine synthase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: L-serine-phosphatidylethanolamine phosphatidyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.30866 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRRGERRDAG GPRPESPVPA GRASLEEPPD GPSAGQATGP GEGRRSTESE VYDDGTNTFF WRAHTLTVLF ILTCTLGYVT LLEETPQDT AYNTKRGIVA SILVFLCFGV TQAKDGPFSR PHPAYWRFWL CVSVVYELFL IFILFQTVQD GRQFLKYVDP K LGVPLPER ...String: MRRGERRDAG GPRPESPVPA GRASLEEPPD GPSAGQATGP GEGRRSTESE VYDDGTNTFF WRAHTLTVLF ILTCTLGYVT LLEETPQDT AYNTKRGIVA SILVFLCFGV TQAKDGPFSR PHPAYWRFWL CVSVVYELFL IFILFQTVQD GRQFLKYVDP K LGVPLPER DYGGNCLIYD PDNETDPFHN IWDKLDGFVP AHFLGWYLKT LMIRDWWMCM IISVMFEFLE YSLEHQLPNF SE CWWDHWI MDVLVCNGLG IYCGMKTLEW LSLKTYKWQG LWNIPTYKGK MKRIAFQFTP YSWVRFEWKP ASSLRRWLAV CGI ILVFLL AELNTFYLKF VLWMPPEHYL VLLRLVFFVN VGGVAMREIY DFMDDPKPHK KLGPQAWLVA AITATELLIV VKYD PHTLT LSLPFYISQC WTLGSVLALT WTVWRFFLRD ITLRYKETRW QKWQNKDDQG STVGNGDQHP LGLDEDLLGP GVAEG EGAP TPN UniProtKB: Phosphatidylserine synthase 2 |
-Macromolecule #2: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phospho...
| Macromolecule | Name: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine type: ligand / ID: 2 / Number of copies: 4 / Formula: P5S |
|---|---|
| Molecular weight | Theoretical: 792.075 Da |
| Chemical component information |  ChemComp-P5S: |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22la...
| Macromolecule | Name: (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate type: ligand / ID: 4 / Number of copies: 2 / Formula: RXY |
|---|---|
| Molecular weight | Theoretical: 715.981 Da |
| Chemical component information | 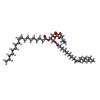 ChemComp-RXY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 14 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































