[English] 日本語
 Yorodumi
Yorodumi- EMDB-48627: Human IMPDH2 mutant - S160del, treated with GTP, ATP, IMP, and NA... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human IMPDH2 mutant - S160del, treated with GTP, ATP, IMP, and NAD+; tetramer reconstruction | |||||||||
 Map data Map data | Output from Phenix density modification, with blur_by_resolution_factor=5, used for model refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dehydrogenase / purine biosynthesis / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology information'de novo' XMP biosynthetic process / Purine ribonucleoside monophosphate biosynthesis / lymphocyte proliferation / IMP dehydrogenase / IMP dehydrogenase activity / GMP biosynthetic process / Azathioprine ADME / peroxisomal membrane / GTP biosynthetic process / cellular response to interleukin-4 ...'de novo' XMP biosynthetic process / Purine ribonucleoside monophosphate biosynthesis / lymphocyte proliferation / IMP dehydrogenase / IMP dehydrogenase activity / GMP biosynthetic process / Azathioprine ADME / peroxisomal membrane / GTP biosynthetic process / cellular response to interleukin-4 / circadian rhythm / secretory granule lumen / ficolin-1-rich granule lumen / Potential therapeutics for SARS / nucleotide binding / Neutrophil degranulation / DNA binding / RNA binding / extracellular exosome / extracellular region / metal ion binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | O'Neill AG / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Overexpression of pathogenic human IMPDH2 variant in Xenopus tropicalis disrupts somitogenesis Authors: O'Neill AG / McCartney ME / Wheeler GM / Patel JH / Kollman JM / Wills AE | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48627.map.gz emd_48627.map.gz | 3.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48627-v30.xml emd-48627-v30.xml emd-48627.xml emd-48627.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48627_fsc.xml emd_48627_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_48627.png emd_48627.png | 50.4 KB | ||
| Masks |  emd_48627_msk_1.map emd_48627_msk_1.map emd_48627_msk_2.map emd_48627_msk_2.map | 103 MB 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-48627.cif.gz emd-48627.cif.gz | 6.6 KB | ||
| Others |  emd_48627_additional_1.map.gz emd_48627_additional_1.map.gz emd_48627_additional_2.map.gz emd_48627_additional_2.map.gz emd_48627_half_map_1.map.gz emd_48627_half_map_1.map.gz emd_48627_half_map_2.map.gz emd_48627_half_map_2.map.gz | 97.2 MB 51.4 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48627 http://ftp.pdbj.org/pub/emdb/structures/EMD-48627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48627 | HTTPS FTP |
-Related structure data
| Related structure data |  9mubMC  9mucC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_48627.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48627.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Output from Phenix density modification, with blur_by_resolution_factor=5, used for model refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.885 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_48627_msk_1.map emd_48627_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_48627_msk_2.map emd_48627_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map from cryoSPARC NU refinement
| File | emd_48627_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from cryoSPARC NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map from cryoSPARC NU refinement
| File | emd_48627_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from cryoSPARC NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A from cryoSPARC NU refinement
| File | emd_48627_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from cryoSPARC NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B from cryoSPARC NU refinement
| File | emd_48627_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from cryoSPARC NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Inosine 5'-monophosphate dehydrogenase 2 - S160del mutant, bound ...
| Entire | Name: Inosine 5'-monophosphate dehydrogenase 2 - S160del mutant, bound to GTP, ATP, IMP, and NAD+ |
|---|---|
| Components |
|
-Supramolecule #1: Inosine 5'-monophosphate dehydrogenase 2 - S160del mutant, bound ...
| Supramolecule | Name: Inosine 5'-monophosphate dehydrogenase 2 - S160del mutant, bound to GTP, ATP, IMP, and NAD+ type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: 5 uM enzyme was mixed with 1 mM ATP, 1 mM MgCl2, 20mM GTP, 1 mM IMP, and 300 uM NAD+. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Inosine-5'-monophosphate dehydrogenase 2
| Macromolecule | Name: Inosine-5'-monophosphate dehydrogenase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: IMP dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.393422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISRDID ...String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISRDID FLKEEEHDCF LEEIMTKRED LVVAPAGITL KEANEILQRS KKGKLPIVNE DDELVAIIAR TDLKKNRDYP LA SKDAKKQ LLCGAAIGTH EDDKYRLDLL AQAGVDVVVL DSSQGNSIFQ INMIKYIKDK YPNLQVIGGN VVTAAQAKNL IDA GVDALR VGMGSGSICI TQEVLACGRP QATAVYKVSE YARRFGVPVI ADGGIQNVGH IAKALALGAS TVMMGSLLAA TTEA PGEYF FSDGIRLKKY RGMGSLDAMD KHLSSQNRYF SEADKIKVAQ GVSGAVQDKG SIHKFVPYLI AGIQHSCQDI GAKSL TQVR AMMYSGELKF EKRTSSAQVE GGVHSLHSYE KRLF UniProtKB: Inosine-5'-monophosphate dehydrogenase 2 |
-Macromolecule #2: INOSINIC ACID
| Macromolecule | Name: INOSINIC ACID / type: ligand / ID: 2 / Number of copies: 4 / Formula: IMP |
|---|---|
| Molecular weight | Theoretical: 348.206 Da |
| Chemical component information | 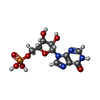 ChemComp-I: |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2630 / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 45000 |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)






































































