[English] 日本語
 Yorodumi
Yorodumi- EMDB-47571: Fully human monoclonal antibody targeting the cysteine-rich subst... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 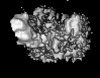 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fully human monoclonal antibody targeting the cysteine-rich substrate-interacting region of ADAM17 on cancer cells. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / complex / ONCOPROTEIN / ANTITUMOR PROTEIN / ANTITUMOR PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology | Domain of unknown function DUF3850 / Domain of unknown function (DUF3850) / ASCH / ASCH domain / PUA-like superfamily / DUF3850 domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Saha N / De La Cruz MJ / Goldgur Y / Nikolov DB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biomed Pharmacother / Year: 2024 Journal: Biomed Pharmacother / Year: 2024Title: Fully human monoclonal antibody targeting the cysteine-rich substrate-interacting region of ADAM17 on cancer cells. Authors: Nayanendu Saha / Sang Gyu Lee / Eeva-Christine Brockmann / M Jason de la Cruz / Yehuda Goldgur / Rachelle P Mendoza / Elisa de Stanchina / Tanzy M Love / Josh Marvald / Yan Xu / Kai Xu / ...Authors: Nayanendu Saha / Sang Gyu Lee / Eeva-Christine Brockmann / M Jason de la Cruz / Yehuda Goldgur / Rachelle P Mendoza / Elisa de Stanchina / Tanzy M Love / Josh Marvald / Yan Xu / Kai Xu / Juha P Himanen / Urpo Lamminmäki / Darren Veach / Dimitar B Nikolov /  Abstract: ADAM17 sheds EGFR/erbB ligands and triggers oncogenic pathways that lead to the progression of solid tumors. We targeted the ADAM17 disintegrin and cysteine rich domain region (D+C) to generate a ...ADAM17 sheds EGFR/erbB ligands and triggers oncogenic pathways that lead to the progression of solid tumors. We targeted the ADAM17 disintegrin and cysteine rich domain region (D+C) to generate a panel of single-chain antibody fragments (scFvs) that selectively bind to the D or C domains of ADAM17, but not of ADAM10 or ADAM19. From the panel, we selected one scFv, referred to as C12, based on its high binding affinity towards the target, and re-formatted it to a full IgG for further studies. High-resolution cryo-electron microscopy studies documented that the mAb binds to the ADAM17 C-domain that in ADAM proteases, notably ADAM10 and ADAM17, is known to impart substrate-specificity. The C12 mAb significantly inhibited EGFR phosphorylation in cancer cell lines by hindering the cleavage of EGFR ligands tethered to the cell surface. This inhibition provides a mechanism for potential anti-tumor effects, and indeed C12 diminished the viability of a variety of EGFR-expressing cancer cell lines. Cell-based ELISA studies revealed that C12 preferentially bound to activated ADAM17 present on tumor cells, as compared to the autoinhibited ADAM17 that is the predominant form on HEK293 and other non-tumor cells. C12 also exhibited tumor growth inhibition in an ovarian cancer xenograft mouse model. Consistent with its selective tumor cell binding in vitro, radioimmuno PET (positron emission tomography) imaging with Zr-DFO-C12 in mouse xenograft models confirmed tumoral accumulation of the C12 mAb. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47571.map.gz emd_47571.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47571-v30.xml emd-47571-v30.xml emd-47571.xml emd-47571.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47571_fsc.xml emd_47571_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_47571.png emd_47571.png | 89.7 KB | ||
| Filedesc metadata |  emd-47571.cif.gz emd-47571.cif.gz | 5.6 KB | ||
| Others |  emd_47571_half_map_1.map.gz emd_47571_half_map_1.map.gz emd_47571_half_map_2.map.gz emd_47571_half_map_2.map.gz | 199.8 MB 199.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47571 http://ftp.pdbj.org/pub/emdb/structures/EMD-47571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47571 | HTTPS FTP |
-Related structure data
| Related structure data |  9e6kMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47571.map.gz / Format: CCP4 / Size: 4.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47571.map.gz / Format: CCP4 / Size: 4.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8255 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_47571_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_47571_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of ADAM17 disintegrin-cysteine rich domain
| Entire | Name: Complex of ADAM17 disintegrin-cysteine rich domain |
|---|---|
| Components |
|
-Supramolecule #1: Complex of ADAM17 disintegrin-cysteine rich domain
| Supramolecule | Name: Complex of ADAM17 disintegrin-cysteine rich domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: heavy chain of monoclonal antibody C12
| Macromolecule | Name: heavy chain of monoclonal antibody C12 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.02983 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: EVQLLESGGG LVQPGGSLRL SCAASGFTFS GYWMHWVRQA PGKGLEWVSR ITYNGTTDYA DSVKGRFTIS RDNSKNTLYL QMNSLRAED TAVYYCARGW LDVWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL T SGVHTFPA ...String: EVQLLESGGG LVQPGGSLRL SCAASGFTFS GYWMHWVRQA PGKGLEWVSR ITYNGTTDYA DSVKGRFTIS RDNSKNTLYL QMNSLRAED TAVYYCARGW LDVWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL T SGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSC |
-Macromolecule #2: light chain monoclonal antibody C12
| Macromolecule | Name: light chain monoclonal antibody C12 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.439961 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSNLAWYQQK PGQAPRLLIY GASSRATGVP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQSYSLPWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSNLAWYQQK PGQAPRLLIY GASSRATGVP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQSYSLPWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #3: Disintegrin and metalloproteinase domain-containing protein 17
| Macromolecule | Name: Disintegrin and metalloproteinase domain-containing protein 17 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.4.24.86 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.579383 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: AQKKCQEAIN ATCKGVSYCT GNSSECPPPG NAEDDTVCLD LGKCKDGKCI PFCEREQQLE SCACNETDNS CKVCCRDLSG RCVPYVDAE QKNLFLRKGK PCTVGFCDMN GKCEKR UniProtKB: DUF3850 domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)






































