+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4754 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Escherichia coli AGPase in complex with FBP. | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ADP-glucose pyrophosphorilase Complex with FBP activator / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucose-1-phosphate adenylyltransferase complex / glucose-1-phosphate adenylyltransferase / glucose-1-phosphate adenylyltransferase activity / glycogen biosynthetic process / AMP binding / protein homotetramerization / magnesium ion binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Cifuente JO / Comino N | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM Authors: Cifuente JO / Comino N / D'Angelo C / Marina A / Gil-Carton D / Albesa-Jove D / Guerin ME | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4754.map.gz emd_4754.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4754-v30.xml emd-4754-v30.xml emd-4754.xml emd-4754.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4754_fsc.xml emd_4754_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4754.png emd_4754.png | 128.4 KB | ||
| Filedesc metadata |  emd-4754.cif.gz emd-4754.cif.gz | 6.9 KB | ||
| Others |  emd_4754_additional_1.map.gz emd_4754_additional_1.map.gz emd_4754_additional_2.map.gz emd_4754_additional_2.map.gz emd_4754_additional_3.map.gz emd_4754_additional_3.map.gz | 28.2 MB 28.2 MB 15.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4754 http://ftp.pdbj.org/pub/emdb/structures/EMD-4754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4754 | HTTPS FTP |
-Validation report
| Summary document |  emd_4754_validation.pdf.gz emd_4754_validation.pdf.gz | 531.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4754_full_validation.pdf.gz emd_4754_full_validation.pdf.gz | 531 KB | Display | |
| Data in XML |  emd_4754_validation.xml.gz emd_4754_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_4754_validation.cif.gz emd_4754_validation.cif.gz | 12.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4754 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4754 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4754 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4754 | HTTPS FTP |
-Related structure data
| Related structure data |  6r8bMC  4761C  6r8uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4754.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4754.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: None
| File | emd_4754_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: None
| File | emd_4754_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: None
| File | emd_4754_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ADP.glucose pyrophosphorylase in complex with the activator FBP
| Entire | Name: ADP.glucose pyrophosphorylase in complex with the activator FBP |
|---|---|
| Components |
|
-Supramolecule #1: ADP.glucose pyrophosphorylase in complex with the activator FBP
| Supramolecule | Name: ADP.glucose pyrophosphorylase in complex with the activator FBP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Homotetrameric enzyme |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 194 KDa |
-Macromolecule #1: Glucose-1-phosphate adenylyltransferase
| Macromolecule | Name: Glucose-1-phosphate adenylyltransferase / type: protein_or_peptide / ID: 1 / Details: 433 Fructose 1,6-Bi-Phosphate / Number of copies: 4 / Enantiomer: LEVO / EC number: glucose-1-phosphate adenylyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.75859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVSLEKNDHL MLARQLPLKS VALILAGGRG TRLKDLTNKR AKPAVHFGGK FRIIDFALSN CINSGIRRMG VITQYQSHTL VQHIQRGWS FFNEEMNEFV DLLPAQQRMK GENWYRGTAD AVTQNLDIIR RYKAEYVVIL AGDHIYKQDY SRMLIDHVEK G ARCTVACM ...String: MVSLEKNDHL MLARQLPLKS VALILAGGRG TRLKDLTNKR AKPAVHFGGK FRIIDFALSN CINSGIRRMG VITQYQSHTL VQHIQRGWS FFNEEMNEFV DLLPAQQRMK GENWYRGTAD AVTQNLDIIR RYKAEYVVIL AGDHIYKQDY SRMLIDHVEK G ARCTVACM PVPIEEASAF GVMAVDENDK IIEFVEKPAN PPSMPNDPSK SLASMGIYVF DADYLYELLE EDDRDENSSH DF GKDLIPK ITEAGLAYAH PFPLSCVQSD PDAEPYWRDV GTLEAYWKAN LDLASVVPEL DMYDRNWPIR TYNESLPPAK FVQ DRSGSH GMTLNSLVSG GCVISGSVVV QSVLFSRVRV NSFCNIDSAV LLPEVWVGRS CRLRRCVIDR ACVIPEGMVI GENA EEDAR RFYRSEEGIV LVTREMLRKL GHKQER UniProtKB: Glucose-1-phosphate adenylyltransferase |
-Macromolecule #2: 1,6-di-O-phosphono-beta-D-fructofuranose
| Macromolecule | Name: 1,6-di-O-phosphono-beta-D-fructofuranose / type: ligand / ID: 2 / Number of copies: 4 / Formula: FBP |
|---|---|
| Molecular weight | Theoretical: 340.116 Da |
| Chemical component information | 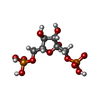 ChemComp-FBP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK II | ||||||||
| Details | Sample monodisperse on graphene oxide home made grids |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Max: 80.0 K |
| Details | Titan Krios I - Ebic - Diamond Light Source |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)