[English] 日本語
 Yorodumi
Yorodumi- EMDB-47148: CRYO-EM STRUCTURE OF SACCHAROMYCES CEREVISIAE RAT1-RAI1-RTT103 COMPLEX -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYO-EM STRUCTURE OF SACCHAROMYCES CEREVISIAE RAT1-RAI1-RTT103 COMPLEX | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Nuclease / Complex / Transcription termination / TRANSCRIPTION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase II termination complex / sno(s)RNA processing / positive regulation of termination of RNA polymerase II transcription / RNA NAD+-cap (NAD+-forming) hydrolase activity / termination of RNA polymerase II transcription, poly(A)-coupled / Las1 complex / termination of RNA polymerase II transcription, exosome-dependent / Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA / Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / phosphodiesterase decapping endonuclease activity ...RNA polymerase II termination complex / sno(s)RNA processing / positive regulation of termination of RNA polymerase II transcription / RNA NAD+-cap (NAD+-forming) hydrolase activity / termination of RNA polymerase II transcription, poly(A)-coupled / Las1 complex / termination of RNA polymerase II transcription, exosome-dependent / Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA / Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / phosphodiesterase decapping endonuclease activity / deadenylation-independent decapping of nuclear-transcribed mRNA / mRNA 5'-diphosphatase activity / RNA polymerase II C-terminal domain phosphoserine binding / NAD-cap decapping / nuclear polyadenylation-dependent rRNA catabolic process / 5'-3' RNA exonuclease activity / nuclear mRNA surveillance / transposable element silencing / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / mRNA 3'-end processing / RNA polymerase II transcribes snRNA genes / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / RNA polymerase II complex binding / nuclear-transcribed mRNA catabolic process / negative regulation of transcription elongation by RNA polymerase II / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / enzyme regulator activity / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / mRNA splicing, via spliceosome / mRNA processing / rRNA processing / site of double-strand break / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / rRNA binding / nucleotide binding / mRNA binding / chromatin / mitochondrion / DNA binding / RNA binding / metal ion binding / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
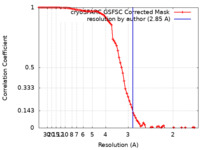
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | ||||||||||||
 Authors Authors | Chu HF / Tong L | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Molecular basis for the interaction between Saccharomyces cerevisiae Rtt103 and the Rat1-Rai1 complex. Authors: Hsu-Feng Chu / Liang Tong /  Abstract: The Rat1 5'-3' exoribonuclease together with its partner Rai1 have important roles in Saccharomyces cerevisiae RNA polymerase II transcription termination. Rtt103 copurifies with Rat1-Rai1 in S. ...The Rat1 5'-3' exoribonuclease together with its partner Rai1 have important roles in Saccharomyces cerevisiae RNA polymerase II transcription termination. Rtt103 copurifies with Rat1-Rai1 in S. cerevisiae, but its mechanism of interaction with them is not known. We report here the cryo-EM structure of the S. cerevisiae Rat1-Rai1-Rtt103 ternary complex at 2.9 Å resolution. We found that a short segment of Rtt103 is in close contact with Rai1, while the rest of Rtt103, including its RNA polymerase II C-terminal domain interaction domain, shows no interactions with Rai1 or Rat1. This is in contrast to the observations on the Komagataella phaffii Rat1-Rai1-Rtt103 complex, where only the RNA polymerase II C-terminal domain interaction domain of Rtt103 has contacts with Rai1. Our structure reveals that S. cerevisiae Rtt103 Pro261 and Tyr263 have important contacts with Rai1, and we show that the P261G/Y263A mutation of Rtt103 blocks the interaction with Rat1-Rai1. Our structure suggests that, in yeast, this segment of Rtt103, which we have named the Rai1 interaction segment, likely helps the recruitment of Rat1-Rai1 to RNA polymerase II for termination. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47148.map.gz emd_47148.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47148-v30.xml emd-47148-v30.xml emd-47148.xml emd-47148.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47148_fsc.xml emd_47148_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_47148.png emd_47148.png | 104.4 KB | ||
| Filedesc metadata |  emd-47148.cif.gz emd-47148.cif.gz | 7.4 KB | ||
| Others |  emd_47148_half_map_1.map.gz emd_47148_half_map_1.map.gz emd_47148_half_map_2.map.gz emd_47148_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47148 http://ftp.pdbj.org/pub/emdb/structures/EMD-47148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47148 | HTTPS FTP |
-Validation report
| Summary document |  emd_47148_validation.pdf.gz emd_47148_validation.pdf.gz | 824.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_47148_full_validation.pdf.gz emd_47148_full_validation.pdf.gz | 823.7 KB | Display | |
| Data in XML |  emd_47148_validation.xml.gz emd_47148_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_47148_validation.cif.gz emd_47148_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47148 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47148 | HTTPS FTP |
-Related structure data
| Related structure data |  9dsoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47148.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47148.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_47148_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_47148_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rat1-Rai1-Rtt103 complex
| Entire | Name: Rat1-Rai1-Rtt103 complex |
|---|---|
| Components |
|
-Supramolecule #1: Rat1-Rai1-Rtt103 complex
| Supramolecule | Name: Rat1-Rai1-Rtt103 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 5'-3' exoribonuclease 2
| Macromolecule | Name: 5'-3' exoribonuclease 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 114.204031 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGVPSFFRWL SRKYPKIISP VLEEQPQIVD GVILPLDYSA SNPNGELDNL YLDMNGIVHP CSHPENKPPP ETEDEMLLAV FEYTNRVLN MARPRKVLVM AVDGVAPRAK MNQQRARRFR SARDAQIENE AREEIMRQRE EVGEIIDDAV RNKKTWDSNA I TPGTPFMD ...String: MGVPSFFRWL SRKYPKIISP VLEEQPQIVD GVILPLDYSA SNPNGELDNL YLDMNGIVHP CSHPENKPPP ETEDEMLLAV FEYTNRVLN MARPRKVLVM AVDGVAPRAK MNQQRARRFR SARDAQIENE AREEIMRQRE EVGEIIDDAV RNKKTWDSNA I TPGTPFMD KLAAALRYWT AFKLATDPGW KNLQVIISDA TVPGEGEHKI MNFIRSQRAD PEYNPNTTHC IYGLDADLIF LG LATHEPH FKILREDVFA QDNRKRNNLK DTINMTEEEK QFLQKQNSEQ PFLWLHINVL REYLSAELWV PGLPFTFDLE RAI DDWVFM CFFCGNDFLP HLPCLDVREN SIDILLDIWK VVLPKLKTYM TCDGVLNLPS VETLLQHLGS REGDIFKTRH IQEA RKKEA FERRKAQKNM SKGQDRHPTV ATEQLQMYDT QGNLAKGSWN LTTSDMVRLK KELMLANEGN EEAIAKVKQQ SDKNN ELMK DISKEEIDDA VSKANKTNFN LAEVMKQKII NKKHRLEKDN EEEEIAKDSK KVKTEKAESE CDLDAEIKDE IVADVN DRE NSETTEVSRD SPVHSTVNVS EGPKNGVFDT DEFVKLFEPG YHERYYTAKF HVTPQDIEQL RKDMVKCYIE GVAWVLM YY YQGCASWNWF YPYHYAPLAT DFHGFSHLEI KFEEGTPFLP YEQLMSVLPA ASGHALPKIF RSLMSEPDSE IIDFYPEE F PIDMNGKKMS WQGIALLPFI DQDRLLTAVR AQYPLLSDAE RARNIRGEPV LLISNKNANY ERFSKKLYSK ENNNNNVVV KFQHFKSGLS GIVSKDVEGF ELNGKIVCPI QGGSLPNLST TLILKMSYRL IPLPSRNKSI ILNGFIPSEP VLTAYDLDSI MYKYNNQNY SRRWNFGNDL KQNIVPVGPK GITQYKPRTG GYRAFFYFAE LSRNNVQPAH NYGRNSYNSQ PGFNNSRYDG G NNNYRQNS NYRNNKYSGN RNSLEHHHHH H UniProtKB: 5'-3' exoribonuclease 2 |
-Macromolecule #2: Decapping nuclease RAI1
| Macromolecule | Name: Decapping nuclease RAI1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.571445 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGVSANLFVK QRGSTTALKQ PKEIGFYSRT KDEEYLISDD TNLNYYYLPD AELDRKLDLS SGFQKFKDYY KDFEDRCSLR GLLETIESS ERHKGKKINA DIITFRGIAR KLISCAFDSP SFNTVDLRIV SFNGQLFIKE VPEAVNAAKA SSATEAGRNI N QDLNVFTG ...String: MGVSANLFVK QRGSTTALKQ PKEIGFYSRT KDEEYLISDD TNLNYYYLPD AELDRKLDLS SGFQKFKDYY KDFEDRCSLR GLLETIESS ERHKGKKINA DIITFRGIAR KLISCAFDSP SFNTVDLRIV SFNGQLFIKE VPEAVNAAKA SSATEAGRNI N QDLNVFTG YKFETLATLS NPLQYTPREV IEKRTKRIVS HGDEYISVVR TGVGNCKLIL GAEVDCIFDF KENGRDNLKH YA ELKCTQQ VANISDTHKF ERKLFRTWLQ CFLVGIPRII YGFKDDHYVL KTVEEFSTEE VPVLLKNNNP QVGSACLEAI KWY GLLTEW LLKMIPRDED PHSQIRAFKL VFENNHLRLS EIEESDEEYS GLIDGEHILS NGFKEWRKSL K UniProtKB: Decapping nuclease RAI1 |
-Macromolecule #3: Regulator of Ty1 transposition protein 103
| Macromolecule | Name: Regulator of Ty1 transposition protein 103 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.725473 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MPFSSEQFTT KLNTLEDSQE SISSASKWLL LQYRDAPKVA EMWKEYMLRP SVNTRRKLLG LYLMNHVVQ QAKGQKIIQF QDSFGKVAAE VLGRINQEFP RDLKKKLSRV VNILKERNIF SKQVVNDIER SLKTESSPVE A LVLPQKLK ...String: MGSSHHHHHH SSGLVPRGSH MPFSSEQFTT KLNTLEDSQE SISSASKWLL LQYRDAPKVA EMWKEYMLRP SVNTRRKLLG LYLMNHVVQ QAKGQKIIQF QDSFGKVAAE VLGRINQEFP RDLKKKLSRV VNILKERNIF SKQVVNDIER SLKTESSPVE A LVLPQKLK DFAKDYEKLV KMHHNVCAMK MRFDKSSDEL DPSSSVYEEN FKTISKIGNM AKDIINESIL KRESGIHKLQ ST LDDEKRH LDEEQNMLSE IEFVLSAKDP SRLNKNVDED NIIPTYEVGD GDDDDDDGDN DDDDDDDDDD KNYDDRSNDS NYG VTNIST TDKKNEVVEK TDSEHKNSTH NPSDNQFGMK RTHDMIGHDD ANDIPEKKVH LDSKTSEDGT FNSEDGHYEL DIEG HVGAQ TDEGVENSGG VSSSIQDLLS KLAN UniProtKB: Regulator of Ty1 transposition protein 103 |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 14 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: FLEXIBLE FIT |
| Output model |  PDB-9dso: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)