[English] 日本語
 Yorodumi
Yorodumi- EMDB-4713: Negative staining of antibodies cooperative complex formed by hum... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4713 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative staining of antibodies cooperative complex formed by human fab7B10 and fab2C1 bound to the N. meningitidis antigen factor H binding protein (fHbp) | |||||||||
 Map data Map data | Negative staining of cooperative couple of human fab7B10 and fab2C1 bound to N.meningitidis antigen fHbp | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Neisseria meningitidis serogroup B (bacteria) Neisseria meningitidis serogroup B (bacteria) | |||||||||
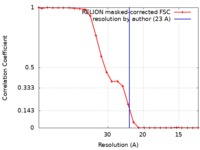
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Peschiera I / Ferlenghi I / Melero R / Liljeroos LJ / Paccagnini E / Giusti F / Carazo JM / Sorzano COS / Scarselli M | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2019 Journal: Commun Biol / Year: 2019Title: Structural basis for cooperativity of human monoclonal antibodies to meningococcal factor H-binding protein. Authors: Ilaria Peschiera / Maria Giuliani / Fabiola Giusti / Roberto Melero / Eugenio Paccagnini / Danilo Donnarumma / Werner Pansegrau / José M Carazo / Carlos O S Sorzano / Maria Scarselli / Vega ...Authors: Ilaria Peschiera / Maria Giuliani / Fabiola Giusti / Roberto Melero / Eugenio Paccagnini / Danilo Donnarumma / Werner Pansegrau / José M Carazo / Carlos O S Sorzano / Maria Scarselli / Vega Masignani / Lassi J Liljeroos / Ilaria Ferlenghi /    Abstract: Monoclonal antibody (mAb) cooperativity is a phenomenon triggered when mAbs couples promote increased bactericidal killing compared to individual partners. Cooperativity has been deeply investigated ...Monoclonal antibody (mAb) cooperativity is a phenomenon triggered when mAbs couples promote increased bactericidal killing compared to individual partners. Cooperativity has been deeply investigated among mAbs elicited by factor H-binding protein (fHbp), a surface-exposed lipoprotein and one of the key antigens included in both serogroup B meningococcus vaccine Bexsero and Trumenba. Here we report the structural and functional characterization of two cooperative mAbs pairs isolated from Bexsero vaccines. The 3D electron microscopy structures of the human mAb-fHbp-mAb cooperative complexes indicate that the angle formed between the antigen binding fragments (fAbs) assume regular angle and that fHbp is able to bind simultaneously and stably the cooperative mAbs pairs and human factor H (fH) in vitro. These findings shed light on molecular basis of the antibody-based mechanism of protection driven by simultaneous recognition of the different epitopes of the fHbp and underline that cooperativity is crucial in vaccine efficacy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4713.map.gz emd_4713.map.gz | 785.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4713-v30.xml emd-4713-v30.xml emd-4713.xml emd-4713.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4713_fsc.xml emd_4713_fsc.xml | 2.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4713.png emd_4713.png | 116 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4713 http://ftp.pdbj.org/pub/emdb/structures/EMD-4713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4713 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4713.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4713.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative staining of cooperative couple of human fab7B10 and fab2C1 bound to N.meningitidis antigen fHbp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex formed by fAb7B10, fAb2C1 and a molecule of fHbp
| Entire | Name: Ternary complex formed by fAb7B10, fAb2C1 and a molecule of fHbp |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex formed by fAb7B10, fAb2C1 and a molecule of fHbp
| Supramolecule | Name: Ternary complex formed by fAb7B10, fAb2C1 and a molecule of fHbp type: complex / ID: 1 / Parent: 0 Details: The cooperative couple of fab was generated using the same CDR region of the cooperative couple of human IgG1 mAb7B10 and mAb2C1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Supramolecule #2: N.meningitidis antigen factor H binding protein (fHbp)
| Supramolecule | Name: N.meningitidis antigen factor H binding protein (fHbp) type: organelle_or_cellular_component / ID: 2 / Parent: 1 Details: Is a surface-exposed lipoprotein expressed at different levels among the strains and it is present as recombinant antigen in both vaccines against meningococcal serogroup B licensed so far |
|---|---|
| Source (natural) | Organism:  Neisseria meningitidis serogroup B (bacteria) Neisseria meningitidis serogroup B (bacteria) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: 1% Uranyl acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)