[English] 日本語
 Yorodumi
Yorodumi- EMDB-4597: T=4 quasi symmetric bacterial microcompartment particle with one ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4597 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T=4 quasi symmetric bacterial microcompartment particle with one missing pentameric EutN unit | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.75 Å | |||||||||
 Authors Authors | Kalnins G | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Authors: Gints Kalnins / Eva-Emilija Cesle / Juris Jansons / Janis Liepins / Anatolij Filimonenko / Kaspars Tars /   Abstract: Bacterial microcompartments (BMCs) are prokaryotic organelles consisting of a protein shell and an encapsulated enzymatic core. BMCs are involved in several biochemical processes, such as choline, ...Bacterial microcompartments (BMCs) are prokaryotic organelles consisting of a protein shell and an encapsulated enzymatic core. BMCs are involved in several biochemical processes, such as choline, glycerol and ethanolamine degradation and carbon fixation. Since non-native enzymes can also be encapsulated in BMCs, an improved understanding of BMC shell assembly and encapsulation processes could be useful for synthetic biology applications. Here we report the isolation and recombinant expression of BMC structural genes from the Klebsiella pneumoniae GRM2 locus, the investigation of mechanisms behind encapsulation of the core enzymes, and the characterization of shell particles by cryo-EM. We conclude that the enzymatic core is encapsulated in a hierarchical manner and that the CutC choline lyase may play a secondary role as an adaptor protein. We also present a cryo-EM structure of a pT = 4 quasi-symmetric icosahedral shell particle at 3.3 Å resolution, and demonstrate variability among the minor shell forms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4597.map.gz emd_4597.map.gz | 97.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4597-v30.xml emd-4597-v30.xml emd-4597.xml emd-4597.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4597_fsc.xml emd_4597_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4597.png emd_4597.png | 70.7 KB | ||
| Others |  emd_4597_half_map_1.map.gz emd_4597_half_map_1.map.gz emd_4597_half_map_2.map.gz emd_4597_half_map_2.map.gz | 98.3 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4597 http://ftp.pdbj.org/pub/emdb/structures/EMD-4597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4597 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4597.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4597.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
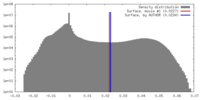
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_4597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
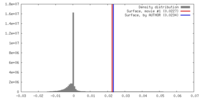
| Density Histograms |
-Half map: #2
| File | emd_4597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
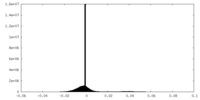
| Density Histograms |
- Sample components
Sample components
-Entire : bacterial microcompartment particles
| Entire | Name: bacterial microcompartment particles |
|---|---|
| Components |
|
-Supramolecule #1: bacterial microcompartment particles
| Supramolecule | Name: bacterial microcompartment particles / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: bacterial microcompartment particle obtained by recombinant expression of pentameric EutN and hexameric cmcC subunits |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) / Strain: MSCL535 Klebsiella pneumoniae (bacteria) / Strain: MSCL535 |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 2.55 MDa |
-Macromolecule #1: cmcD/EutN BMC-P bacterial microcompartment protein
| Macromolecule | Name: cmcD/EutN BMC-P bacterial microcompartment protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MILAKVTGHV VATQKCDELR GSNLLLITRL DDKQQPMKDQ TWVAVDNVGA GMHDIVLAEE YFALNKDRYK AMSVVAIVEK VFRD |
-Macromolecule #2: cmcC/PduA/ccmK BMC-H bacterial microcompartment protein
| Macromolecule | Name: cmcC/PduA/ccmK BMC-H bacterial microcompartment protein type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKEALGLIET KGLVACIEAA DAMCKAANVE LIGYENVGSG LVTAMVKGDV GAVNAAVDSG VEAAKRIGEV VSSRVIARPH NDIEKIAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. | |||||||||
| Details | Sample was purified with gel filtration |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 1316 / Average exposure time: 1.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.4 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)