+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Substrate bound DosP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Heme / DosP / Phosphodiesterase / c-di-GMP / OXYGEN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationcyclic-guanylate-specific phosphodiesterase / regulation of single-species biofilm formation / cyclic-guanylate-specific phosphodiesterase activity / response to oxygen levels / oxygen sensor activity / heme binding / magnesium ion binding / protein homodimerization activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Kumar P / Kober DL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structures of the multi-domain oxygen sensor DosP: remote control of a c-di-GMP phosphodiesterase by a regulatory PAS domain. Authors: Wenbi Wu / Pankaj Kumar / Chad A Brautigam / Shih-Chia Tso / Hamid R Baniasadi / Daniel L Kober / Marie-Alda Gilles-Gonzalez /  Abstract: The heme-based direct oxygen sensor DosP degrades c-di-GMP, a second messenger nearly unique to bacteria. In stationary phase Escherichia coli, DosP is the most abundant c-di-GMP phosphodiesterase. ...The heme-based direct oxygen sensor DosP degrades c-di-GMP, a second messenger nearly unique to bacteria. In stationary phase Escherichia coli, DosP is the most abundant c-di-GMP phosphodiesterase. Ligation of O to a heme-binding PAS domain (hPAS) of the protein enhances the phosphodiesterase through an allosteric mechanism that has remained elusive. We determine six structures of full-length DosP in its aerobic or anaerobic conformations, with or without c-di-GMP. DosP is an elongated dimer with the regulatory heme containing domain and phosphodiesterase separated by nearly 180 Å. In the absence of substrate, regardless of the heme status, DosP presents an equilibrium of two distinct conformations. Binding of substrate induces DosP to adopt a single, ON-state or OFF-state conformation depending on its heme status. Structural and biochemical studies of this multi-domain sensor and its mutants provide insights into signal regulation of second-messenger levels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45746.map.gz emd_45746.map.gz | 50.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45746-v30.xml emd-45746-v30.xml emd-45746.xml emd-45746.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45746.png emd_45746.png | 42.4 KB | ||
| Filedesc metadata |  emd-45746.cif.gz emd-45746.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45746 http://ftp.pdbj.org/pub/emdb/structures/EMD-45746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45746 | HTTPS FTP |
-Related structure data
| Related structure data |  9cmfMC  9bgvC  9bkvC  9cdrC  9ce0C  9cloC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45746.map.gz / Format: CCP4 / Size: 71.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45746.map.gz / Format: CCP4 / Size: 71.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35125 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Dimer of DosP
| Entire | Name: Dimer of DosP |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of DosP
| Supramolecule | Name: Dimer of DosP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Oxygen sensor protein DosP
| Macromolecule | Name: Oxygen sensor protein DosP / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: cyclic-guanylate-specific phosphodiesterase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.202969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIFFPALEQN MMGAVLINEN DEVMFFNPAA EKLWGYKREE VIGNNIDMLI PRDLRPAHPE YIRHNREGGK ARVEGMSREL QLEKKDGSK IWTRFALSKV SAEGKVYYLA LVRDASVEMA QKEQTRQLII AVDHLDRPVI VLDPERHIVQ CNRAFTEMFG Y CISEASGM ...String: GIFFPALEQN MMGAVLINEN DEVMFFNPAA EKLWGYKREE VIGNNIDMLI PRDLRPAHPE YIRHNREGGK ARVEGMSREL QLEKKDGSK IWTRFALSKV SAEGKVYYLA LVRDASVEMA QKEQTRQLII AVDHLDRPVI VLDPERHIVQ CNRAFTEMFG Y CISEASGM QPDTLLNTPE FPADNRIRLQ QLLWKTARDQ DEFLLLTRTG EKIWIKASIS PVYDVLAHLQ NLVMTFSDIT EE RQIRQLE GNILAAMCSS PPFHEMGEII CRNIESVLNE SHVSLFALRN GMPIHWASSS HGAEIQNAQS WSATIRQRDG APA GILQIK TSSGAETSAF IERVADISQH MAALALEQEK SRQHIEQLIQ FDPMTGLPNR NNLHNYLDDL VDKAVSPVVY LIGV DHIQD VIDSLGYAWA DQALLEVVNR FREKLKPDQY LCRIEGTQFV LVSLENDVSN ITQIADELRN VVSKPIMIDD KPFPL TLSI GISYDLGKNR DYLLSTAHNA MDYIRKNGGN GWQFFSPAMN EMVKERLVLG AALKEAISNN QLKLVYQPQI FAETGE LYG IEALARWHDP LHGHVPPSRF IPLAEEIGEI ENIGRWVIAE ACRQLAEWRS QNIHIPALSV NLSALHFRSN QLPNQVS DA MHAWGIDGHQ LTVEITESMM MEHDTEIFKR IQILRDMGVG LSVDDFGTGF SGLSRLVSLP VTEIKIDKSF VDRCLTEK R ILALLEAITS IGQSLNLTVV AEGVETKEQF EMLRKIHCRV IQGYFFSRPL PAEEIPGWMS SVLPLKI UniProtKB: Oxygen sensor protein DosP |
-Macromolecule #2: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 2 / Number of copies: 2 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #3: OXYGEN MOLECULE
| Macromolecule | Name: OXYGEN MOLECULE / type: ligand / ID: 3 / Number of copies: 2 / Formula: OXY |
|---|---|
| Molecular weight | Theoretical: 31.999 Da |
| Chemical component information |  ChemComp-O2: |
-Macromolecule #4: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydr...
| Macromolecule | Name: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one) type: ligand / ID: 4 / Number of copies: 2 / Formula: C2E |
|---|---|
| Molecular weight | Theoretical: 690.411 Da |
| Chemical component information | 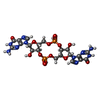 ChemComp-C2E: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.11 Å / Resolution method: FSC 0.143 CUT-OFF Details: This is a composite map created by combining maps generated separately for each half of the protein. The particle count and resolution provided refer to the best map obtained. Number images used: 307809 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4.2) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4.2) |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)