+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | LONP1 stall state bound to substrate and 4 ADPs | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATPase / protease / HYDROLASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
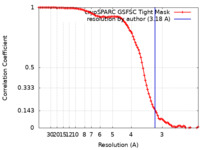
| Method | single particle reconstruction / cryo EM / Resolution: 3.18 Å | |||||||||
 Authors Authors | Mindrebo JT / Lander GC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Structural and mechanistic studies on human LONP1 redefine the hand-over-hand translocation mechanism. Authors: Jeffrey T Mindrebo / Gabriel C Lander /  Abstract: AAA+ enzymes use energy from ATP hydrolysis to remodel diverse cellular targets. Structures of substrate-bound AAA+ complexes suggest that these enzymes employ a conserved hand-over-hand mechanism to ...AAA+ enzymes use energy from ATP hydrolysis to remodel diverse cellular targets. Structures of substrate-bound AAA+ complexes suggest that these enzymes employ a conserved hand-over-hand mechanism to thread substrates through their central pore. However, the fundamental aspects of the mechanisms governing motor function and substrate processing within specific AAA+ families remain unresolved. We used cryo-electron microscopy to structurally interrogate reaction intermediates from in vitro biochemical assays to inform the underlying regulatory mechanisms of the human mitochondrial AAA+ protease, LONP1. Our results demonstrate that substrate binding allosterically regulates proteolytic activity, and that LONP1 can adopt a configuration conducive to substrate translocation even when the ATPases are bound to ADP. These results challenge the conventional understanding of the hand-over-hand translocation mechanism, giving rise to an alternative model that aligns more closely with biochemical and biophysical data on related enzymes like ClpX, ClpA, the 26S proteasome, and Lon protease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45434.map.gz emd_45434.map.gz | 47.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45434-v30.xml emd-45434-v30.xml emd-45434.xml emd-45434.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45434_fsc.xml emd_45434_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_45434.png emd_45434.png | 108.4 KB | ||
| Filedesc metadata |  emd-45434.cif.gz emd-45434.cif.gz | 5.8 KB | ||
| Others |  emd_45434_additional_1.map.gz emd_45434_additional_1.map.gz emd_45434_half_map_1.map.gz emd_45434_half_map_1.map.gz emd_45434_half_map_2.map.gz emd_45434_half_map_2.map.gz | 46 MB 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45434 http://ftp.pdbj.org/pub/emdb/structures/EMD-45434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45434 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45434.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45434.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
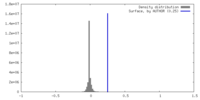
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_45434_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
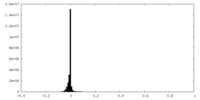
| Density Histograms |
-Half map: Half map B
| File | emd_45434_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
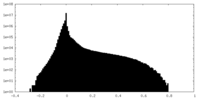
| Density Histograms |
-Half map: Half map A
| File | emd_45434_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
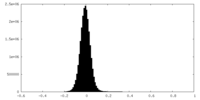
| Density Histograms |
- Sample components
Sample components
-Entire : LONP1 bound to substrate and ADP
| Entire | Name: LONP1 bound to substrate and ADP |
|---|---|
| Components |
|
-Supramolecule #1: LONP1 bound to substrate and ADP
| Supramolecule | Name: LONP1 bound to substrate and ADP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 583.95 KDa |
-Macromolecule #1: Human mitochondrial Lon Protease homolog
| Macromolecule | Name: Human mitochondrial Lon Protease homolog / type: protein_or_peptide / ID: 1 / Details: homohexameric assembly / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGAHMMTI PDVFPHLPLI AITRNPVFPR FIKIIEVKNK KLVELLRRKV RLAQPYVGVF LKRDDSNESD VVESLDEIYH TGTFAQIHEM QDLGDKLRMI VMGHRRVHIS RQLEVEPEEP EAENKHKPRR KSKRGKKEAE DELSARHPAE LAMEPTPELP ...String: MHHHHHHENL YFQGAHMMTI PDVFPHLPLI AITRNPVFPR FIKIIEVKNK KLVELLRRKV RLAQPYVGVF LKRDDSNESD VVESLDEIYH TGTFAQIHEM QDLGDKLRMI VMGHRRVHIS RQLEVEPEEP EAENKHKPRR KSKRGKKEAE DELSARHPAE LAMEPTPELP AEVLMVEVEN VVHEDFQVTE EVKALTAEIV KTIRDIIALN PLYRESVLQM MQAGQRVVDN PIYLSDMGAA LTGAESHELQ DVLEETNIPK RLYKALSLLK KEFELSKLQQ RLGREVEEKI KQTHRKYLLQ EQLKIIKKEL GLEKDDKDAI EEKFRERLKE LVVPKHVMDV VDEELSKLGL LDNHSSEFNV TRNYLDWLTS IPWGKYSNEN LDLARAQAVL EEDHYGMEDV KKRILEFIAV SQLRGSTQGK ILCFYGPPGV GKTSIARSIA RALNREYFRF SVGGMTDVAE IKGHRRTYVG AMPGKIIQCL KKTKTENPLI LIDEVDKIGR GYQGDPSSAL LELLDPEQNA NFLDHYLDVP VDLSKVLFIC TANVTDTIPE PLRDRMEMIN VSGYVAQEKL AIAERYLVPQ ARALCGLDES KAKLSSDVLT LLIKQYCRES GVRNLQKQVE KVLRKSAYKI VSGEAESVEV TPENLQDFVG KPVFTVERMY DVTPPGVVMG LAWTAMGGST LFVETSLRRP QDKDAKGDKD GSLEVTGQLG EVMKESARIA YTFARAFLMQ HAPANDYLVT SHIHLHVPEG ATPKDGPSAG CTIVTALLSL AMGRPVRQNL AMTGEVSLTG KILPVGGIKE KTIAAKRAGV TCIVLPAENK KDFYDLAAFI TEGLEVHFVE HYREIFDIAF PDEQAEALAV ER |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Sample was monodisperse with ~300 particles per image. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3840 pixel / Digitization - Dimensions - Height: 3712 pixel / Number grids imaged: 1 / Number real images: 1944 / Average exposure time: 9.8 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)