[English] 日本語
 Yorodumi
Yorodumi- EMDB-45432: Cryo-EM structure of mouse PI(4,5)P2-bound TRPML1 channel at 2.46... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse PI(4,5)P2-bound TRPML1 channel at 2.46 Angstrom resolution | ||||||||||||
 Map data Map data | structure of mouse PI(4,5)P2-bound TRPML1 channel at 2.46 Angstrom resolution | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | TRPML1 / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationTransferrin endocytosis and recycling / positive regulation of lysosome organization / calcium ion export / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / phagosome maturation / NAADP-sensitive calcium-release channel activity / iron ion transmembrane transporter activity / iron ion transmembrane transport / cellular response to pH / TRP channels ...Transferrin endocytosis and recycling / positive regulation of lysosome organization / calcium ion export / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / phagosome maturation / NAADP-sensitive calcium-release channel activity / iron ion transmembrane transporter activity / iron ion transmembrane transport / cellular response to pH / TRP channels / monoatomic anion channel activity / endosomal transport / intracellular vesicle / sodium channel activity / monoatomic cation transmembrane transport / autophagosome maturation / potassium channel activity / monoatomic cation channel activity / phagocytic cup / release of sequestered calcium ion into cytosol / cellular response to calcium ion / cell projection / calcium channel activity / phagocytic vesicle membrane / late endosome / late endosome membrane / protein homotetramerization / adaptive immune response / lysosome / receptor complex / lysosomal membrane / lipid binding / Golgi apparatus / nucleoplasm / identical protein binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | ||||||||||||
 Authors Authors | Gan N / Jiang Y | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: TRPML1 gating modulation by allosteric mutations and lipids. Authors: Ninghai Gan / Yan Han / Weizhong Zeng / Youxing Jiang /  Abstract: Transient Receptor Potential Mucolipin 1 (TRPML1) is a lysosomal cation channel whose loss-of-function mutations directly cause the lysosomal storage disorder mucolipidosis type IV (MLIV). TRPML1 can ...Transient Receptor Potential Mucolipin 1 (TRPML1) is a lysosomal cation channel whose loss-of-function mutations directly cause the lysosomal storage disorder mucolipidosis type IV (MLIV). TRPML1 can be allosterically regulated by various ligands including natural lipids and small synthetic molecules and the channel undergoes a global movement propagated from ligand-induced local conformational changes upon activation. In this study, we identified a functionally critical residue, Tyr404, at the C-terminus of the S4 helix, whose mutations to tryptophan and alanine yield gain- and loss-of-function channels, respectively. These allosteric mutations mimic the ligand activation or inhibition of the TRPML1 channel without interfering with ligand binding and both mutant channels are susceptible to agonist or antagonist modulation, making them better targets for screening potent TRPML1 activators and inhibitors. We also determined the high-resolution structure of TRPML1 in complex with the PI(4,5)P inhibitor, revealing the structural basis underlying this lipid inhibition. In addition, an endogenous phospholipid likely from sphingomyelin is identified in the PI(4,5)P-bound TRPML1 structure at the same hotspot for agonists and antagonists, providing a plausible structural explanation for the inhibitory effect of sphingomyelin on agonist activation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45432.map.gz emd_45432.map.gz | 84.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45432-v30.xml emd-45432-v30.xml emd-45432.xml emd-45432.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45432.png emd_45432.png | 26.5 KB | ||
| Filedesc metadata |  emd-45432.cif.gz emd-45432.cif.gz | 6.2 KB | ||
| Others |  emd_45432_half_map_1.map.gz emd_45432_half_map_1.map.gz emd_45432_half_map_2.map.gz emd_45432_half_map_2.map.gz | 82.4 MB 82.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45432 http://ftp.pdbj.org/pub/emdb/structures/EMD-45432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45432 | HTTPS FTP |
-Related structure data
| Related structure data |  9cc2MC  9cbzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45432.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45432.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of mouse PI(4,5)P2-bound TRPML1 channel at 2.46 Angstrom resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
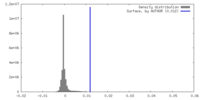
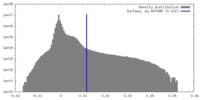
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_45432_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
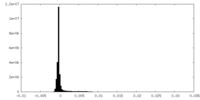
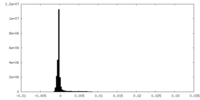
| Density Histograms |
-Half map: Half Map A
| File | emd_45432_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
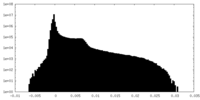
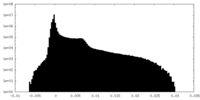
| Density Histograms |
- Sample components
Sample components
-Entire : mouse TRPML1
| Entire | Name: mouse TRPML1 |
|---|---|
| Components |
|
-Supramolecule #1: mouse TRPML1
| Supramolecule | Name: mouse TRPML1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mucolipin-1
| Macromolecule | Name: Mucolipin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.573617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATPAGRRAS ETERLLTPNP GYGTQVGTSP APTTPTEEED LRRRLKYFFM SPCDKFRAKG RKPCKLMLQV VKILVVTVQL ILFGLSNQL VVTFREENTI AFRHLFLLGY SDGSDDTFAA YTQEQLYQAI FYAVDQYLIL PEISLGRYAY VRGGGGPWAN G SALALCQR ...String: MATPAGRRAS ETERLLTPNP GYGTQVGTSP APTTPTEEED LRRRLKYFFM SPCDKFRAKG RKPCKLMLQV VKILVVTVQL ILFGLSNQL VVTFREENTI AFRHLFLLGY SDGSDDTFAA YTQEQLYQAI FYAVDQYLIL PEISLGRYAY VRGGGGPWAN G SALALCQR YYHRGHVDPA NDTFDIDPRV VTDCIQVDPP DRPPDIPSED LDFLDGSASY KNLTLKFHKL INVTIHFQLK TI NLQSLIN NEIPDCYTFS ILITFDNKAH SGRIPIRLET KTHIQECKHP SVSRHGDNSF RLLFDVVVIL TCSLSFLLCA RSL LRGFLL QNEFVVFMWR RRGREISLWE RLEFVNGWYI LLVTSDVLTI SGTVMKIGIE AKNLASYDVC SILLGTSTLL VWVG VIRYL TFFHKYNILI ATLRVALPSV MRFCCCVAVI YLGYCFCGWI VLGPYHVKFR SLSMVSECLF SLINGDDMFV TFAAM QAQQ GHSSLVWLFS QLYLYSFISL FIYMVLSLFI ALITGAYDTI KHPGGTGTEK SELQAYIEQC QDSPTSGKFR RGSGSA CSL FCCCGRDSPE DHSLLVN UniProtKB: Mucolipin-1 |
-Macromolecule #3: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
| Macromolecule | Name: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate type: ligand / ID: 3 / Number of copies: 4 / Formula: PIO |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information |  ChemComp-PIO: |
-Macromolecule #4: sphingomyelin
| Macromolecule | Name: sphingomyelin / type: ligand / ID: 4 / Number of copies: 4 / Formula: FO4 |
|---|---|
| Molecular weight | Theoretical: 814.233 Da |
| Chemical component information |  ChemComp-FO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)