+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Assimilatory NADPH-dependent sulfite reductase minimal dimer | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | sulfite reductase / hemoflavoprotein / oxidoreductase / cryo-EM / FLAVOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsulfite reductase (ferredoxin) activity / assimilatory sulfite reductase (NADPH) / sulfite reductase (NADPH) activity / sulfite reductase complex (NADPH) / sulfate assimilation / hydrogen sulfide biosynthetic process / L-cysteine biosynthetic process / FMN binding / NADP binding / flavin adenine dinucleotide binding ...sulfite reductase (ferredoxin) activity / assimilatory sulfite reductase (NADPH) / sulfite reductase (NADPH) activity / sulfite reductase complex (NADPH) / sulfate assimilation / hydrogen sulfide biosynthetic process / L-cysteine biosynthetic process / FMN binding / NADP binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / heme binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||
 Authors Authors | Ghazi Esfahani B / Walia N / Neselu K / Aragon M / Askenasy I / Wei A / Mendez JH / Stroupe ME | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structure of dimerized assimilatory NADPH-dependent sulfite reductase reveals the minimal interface for diflavin reductase binding. Authors: Behrouz Ghazi Esfahani / Nidhi Walia / Kasahun Neselu / Yashika Garg / Mahira Aragon / Isabel Askenasy / Hui Alex Wei / Joshua H Mendez / M Elizabeth Stroupe /   Abstract: Escherichia coli NADPH-dependent assimilatory sulfite reductase (SiR) reduces sulfite by six electrons to make sulfide for incorporation into sulfur-containing biomolecules. SiR has two subunits: an ...Escherichia coli NADPH-dependent assimilatory sulfite reductase (SiR) reduces sulfite by six electrons to make sulfide for incorporation into sulfur-containing biomolecules. SiR has two subunits: an NADPH, FMN, and FAD-binding diflavin flavoprotein and a siroheme/FeS cluster-containing hemoprotein. The molecular interactions that govern subunit binding have been unknown since the discovery of SiR over 50 years ago because SiR is flexible, thus has been intransigent for traditional high-resolution structural analysis. We use a combination of the chameleon® plunging system with a fluorinated lipid to overcome the challenges of preserving a flexible molecule to determine a 2.78 Å-resolution cryo-EM structure of a minimal heterodimer complex. Chameleon®, combined with the fluorinated lipid, overcomes persistent denaturation at the air-water interface. Using a previously characterized minimal heterodimer reduces the heterogeneity of a structurally heterogeneous complex to a level that we analyze using multi-conformer cryo-EM image analysis algorithms. Here, we report the near-atomic resolution structure of the flavoprotein/hemoprotein complex, revealing how they interact in a minimal interface. Further, we determine the structural elements that discriminate between pairing a hemoprotein with a diflavin reductase, as in the E. coli homolog, or a ferredoxin partner, as in maize (Zea mays). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45359.map.gz emd_45359.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45359-v30.xml emd-45359-v30.xml emd-45359.xml emd-45359.xml | 51 KB 51 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45359_fsc.xml emd_45359_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_45359.png emd_45359.png | 70.9 KB | ||
| Filedesc metadata |  emd-45359.cif.gz emd-45359.cif.gz | 8 KB | ||
| Others |  emd_45359_additional_1.map.gz emd_45359_additional_1.map.gz emd_45359_additional_10.map.gz emd_45359_additional_10.map.gz emd_45359_additional_11.map.gz emd_45359_additional_11.map.gz emd_45359_additional_12.map.gz emd_45359_additional_12.map.gz emd_45359_additional_2.map.gz emd_45359_additional_2.map.gz emd_45359_additional_3.map.gz emd_45359_additional_3.map.gz emd_45359_additional_4.map.gz emd_45359_additional_4.map.gz emd_45359_additional_5.map.gz emd_45359_additional_5.map.gz emd_45359_additional_6.map.gz emd_45359_additional_6.map.gz emd_45359_additional_7.map.gz emd_45359_additional_7.map.gz emd_45359_additional_8.map.gz emd_45359_additional_8.map.gz emd_45359_additional_9.map.gz emd_45359_additional_9.map.gz emd_45359_half_map_1.map.gz emd_45359_half_map_1.map.gz emd_45359_half_map_2.map.gz emd_45359_half_map_2.map.gz | 111.4 MB 48.5 MB 48.6 MB 48.6 MB 131.7 MB 48.7 MB 48.8 MB 48.8 MB 48.7 MB 48.7 MB 48.6 MB 48.5 MB 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45359 http://ftp.pdbj.org/pub/emdb/structures/EMD-45359 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45359 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45359 | HTTPS FTP |
-Related structure data
| Related structure data |  9c91MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45359.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45359.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.765 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: map with the linker
+Additional map: vol 7 gauss z.mrc
+Additional map: vol 8 gauss z.mrc
+Additional map: vol 9 gauss z.mrc
+Additional map: dimer obtained from the SiRFPdelta-SiRHP dataset
+Additional map: vol 0 gauss z.mrc
+Additional map: vol 1 gauss z.mrc
+Additional map: vol 2 gauss z.mrc
+Additional map: vol 3 gauss z.mrc
+Additional map: vol 4 gauss z.mrc
+Additional map: vol 5 gauss z.mrc
+Additional map: vol 6 gauss z.mrc
+Half map: half map A
+Half map: half map B
- Sample components
Sample components
-Entire : sulfite reductase minimal dimer
| Entire | Name: sulfite reductase minimal dimer |
|---|---|
| Components |
|
-Supramolecule #1: sulfite reductase minimal dimer
| Supramolecule | Name: sulfite reductase minimal dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124 KDa |
-Macromolecule #1: Sulfite reductase [NADPH] flavoprotein alpha-component
| Macromolecule | Name: Sulfite reductase [NADPH] flavoprotein alpha-component type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: assimilatory sulfite reductase (NADPH) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.093684 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGSHHHHHH GMASMTGGNN MGRDLYDDDD KDRWGSELEI TIISASQTGN ARRVAEALRD DLLAAKLNVK LVNAGDYKFK QIASEKLLI VVTSTQGCGE PPEEAVALHK FLFSKKAPKL ENTAFAVFSL GDSSYEFFTQ SGKDFDSKLA ELGGERLLDR V DADVEYQA ...String: MGGSHHHHHH GMASMTGGNN MGRDLYDDDD KDRWGSELEI TIISASQTGN ARRVAEALRD DLLAAKLNVK LVNAGDYKFK QIASEKLLI VVTSTQGCGE PPEEAVALHK FLFSKKAPKL ENTAFAVFSL GDSSYEFFTQ SGKDFDSKLA ELGGERLLDR V DADVEYQA AASEWRARVV DALKSRAPVA APSQSVATGA VNEIHTSPYS KDAPLVASLS VNQKITGRNS EKDVRHIEID LG DSGLRYQ PGDALGVWYQ NDPALVKELV ELLWLKGDEP VTVEGKTLPL NEALQWHFEL TVNTANIVEN YATLTRSETL LPL VGDKAK LQHYAATTPI VDMVRFSPAQ LDAEALINLL RPLTPRLYSI ASSQAEVENE VHVTVGVVRY DVEGRARAGG ASSF LADRV EEEGEVRVFI EHNDNFRLPA NPETPVIMIG PGTGIAPFRA FMQQRAADEA PGKNWLFFGN PHFTEDFLYQ VEWQR YVKE GVLTRIDLAW SRDQKEKVYV QDKLREQGAE LWRWINDGAH IYVSGDACRM AKDVEQALLE VIAEFGGMDT EAADEF LSE LRVERRYQRD VY UniProtKB: Sulfite reductase [NADPH] flavoprotein alpha-component |
-Macromolecule #2: Sulfite reductase [NADPH] hemoprotein beta-component
| Macromolecule | Name: Sulfite reductase [NADPH] hemoprotein beta-component / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: assimilatory sulfite reductase (NADPH) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.091102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSEKHPGPLV VEGKLTDAER MKHESNYLRG TIAEDLNDGL TGGFKGDNFL LIRFHGMYQQ DDRDIRAERA EQKLEPRHAM LLRCRLPGG VITTKQWQAI DKFAGENTIY GSIRLTNRQT FQFHGILKKN VKPVHQMLHS VGLDALATAN DMNRNVLCTS N PYESQLHA ...String: MSEKHPGPLV VEGKLTDAER MKHESNYLRG TIAEDLNDGL TGGFKGDNFL LIRFHGMYQQ DDRDIRAERA EQKLEPRHAM LLRCRLPGG VITTKQWQAI DKFAGENTIY GSIRLTNRQT FQFHGILKKN VKPVHQMLHS VGLDALATAN DMNRNVLCTS N PYESQLHA EAYEWAKKIS EHLLPRTRAY AEIWLDQEKV ATTDEEPILG QTYLPRKFKT TVVIPPQNDI DLHANDMNFV AI AENGKLV GFNLLVGGGL SIEHGNKKTY ARTASEFGYL PLEHTLAVAE AVVTTQRDWG NRTDRKNAKT KYTLERVGVE TFK AEVERR AGIKFEPIRP YEFTGRGDRI GWVKGIDDNW HLTLFIENGR ILDYPARPLK TGLLEIAKIH KGDFRITANQ NLII AGVPE SEKAKIEKIA KESGLMNAVT PQRENSMACV SFPTCPLAMA EAERFLPSFI DNIDNLMAKH GVSDEHIVMR VTGCP NGCG RAMLAEVGLV GKAPGRYNLH LGGNRIGTRI PRMYKENITE PEILASLDEL IGRWAKEREA GEGFGDFTVR AGIIRP VLD PARDLWD UniProtKB: Sulfite reductase [NADPH] hemoprotein beta-component |
-Macromolecule #3: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 1 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #4: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #5: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #6: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #8: SIROHEME
| Macromolecule | Name: SIROHEME / type: ligand / ID: 8 / Number of copies: 1 / Formula: SRM |
|---|---|
| Molecular weight | Theoretical: 916.661 Da |
| Chemical component information | 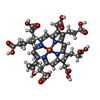 ChemComp-SRM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 6.8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON APOLLO (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































































































































