+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PqqU with ligand PQQ | |||||||||||||||
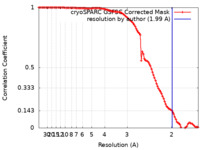
 Map data Map data | Cryo-EM structure of E. coli PqqU with ligand PQQ at 1.99 A global resolution. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | TonD-dependent / outer membrane / transporter / PQQ uptake / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsiderophore transmembrane transport / intracellular monoatomic cation homeostasis / siderophore uptake transmembrane transporter activity / transmembrane transporter complex / cell outer membrane / signaling receptor activity / membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.99 Å | |||||||||||||||
 Authors Authors | Munder F / Venugopal H / Grinter R | |||||||||||||||
| Funding support |  Australia, 4 items Australia, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: High-affinity PQQ import is widespread in Gram-negative bacteria. Authors: Fabian Munder / Marcos Voutsinos / Klaus Hantke / Hari Venugopal / Rhys Grinter /   Abstract: Pyrroloquinoline quinone (PQQ) is a soluble redox cofactor used by diverse bacteria. Many Gram-negative bacteria that encode PQQ-dependent enzymes do not produce it and instead obtain it from the ...Pyrroloquinoline quinone (PQQ) is a soluble redox cofactor used by diverse bacteria. Many Gram-negative bacteria that encode PQQ-dependent enzymes do not produce it and instead obtain it from the environment. To achieve this, uses the TonB-dependent transporter PqqU as a high-affinity PQQ importer. Here, we show that PqqU binds PQQ with high affinity and determine the high-resolution structure of the PqqU-PQQ complex, revealing that PqqU undergoes conformational changes in PQQ binding to capture the cofactor in an internal cavity. We show that these conformational changes preclude the binding of a bacteriophage, which targets PqqU as a cell surface receptor. Guided by the PqqU-PQQ structure, we identify amino acids essential for PQQ import and leverage this information to map the presence of PqqU across Gram-negative bacteria. This reveals that PqqU is encoded by Gram-negative bacteria from at least 22 phyla occupying diverse habitats, indicating that PQQ is an important cofactor for bacteria that adopt diverse lifestyles and metabolic strategies. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45192.map.gz emd_45192.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45192-v30.xml emd-45192-v30.xml emd-45192.xml emd-45192.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45192_fsc.xml emd_45192_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_45192.png emd_45192.png | 189.4 KB | ||
| Filedesc metadata |  emd-45192.cif.gz emd-45192.cif.gz | 7.1 KB | ||
| Others |  emd_45192_half_map_1.map.gz emd_45192_half_map_1.map.gz emd_45192_half_map_2.map.gz emd_45192_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45192 http://ftp.pdbj.org/pub/emdb/structures/EMD-45192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45192 | HTTPS FTP |
-Related structure data
| Related structure data |  9c4oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45192.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45192.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of E. coli PqqU with ligand PQQ at 1.99 A global resolution. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map B of PqqU with ligand PQQ.
| File | emd_45192_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B of PqqU with ligand PQQ. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A of PqqU with ligand PQQ.
| File | emd_45192_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A of PqqU with ligand PQQ. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of PqqU with PQQ
| Entire | Name: Complex of PqqU with PQQ |
|---|---|
| Components |
|
-Supramolecule #1: Complex of PqqU with PQQ
| Supramolecule | Name: Complex of PqqU with PQQ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: TonB-dependent outer membrane transporter PqqU in complex with its substrate PQQ. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78 KDa |
-Macromolecule #1: Pyrroloquinoline quinone transporter
| Macromolecule | Name: Pyrroloquinoline quinone transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.401164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNHHHHHHH HHHENLYFQG AMDIGINSDP ADEQTMIVSA APQVVSELDT PAAVSVVDGE EMRLATPRIN LSESLTGVPG LQVQNRQNY AQDLQLSIRG FGSRSTYGIR GIRLYVDGIP ATMPDGQGQT SNIDLSSVQN VEVLRGPFSA LYGNASGGVM N VTTQTGQQ ...String: MSNHHHHHHH HHHENLYFQG AMDIGINSDP ADEQTMIVSA APQVVSELDT PAAVSVVDGE EMRLATPRIN LSESLTGVPG LQVQNRQNY AQDLQLSIRG FGSRSTYGIR GIRLYVDGIP ATMPDGQGQT SNIDLSSVQN VEVLRGPFSA LYGNASGGVM N VTTQTGQQ PPTIEASSYY GSFGSWRYGL KATGATGDGT QPGDVDYTVS TTRFTTHGYR DHSGAQKNLA NAKLGVRIDE AS KLSLIFN SVDIKADDPG GLTKAEWKAN PQQAPRAEQY DTRKTIKQTQ AGLRYERSLS SRDDMSVMMY AGERETTQYQ SIP MAPQLN PSHAGGVITL QRHYQGIDSR WTHRGELGVP VTFTTGLNYE NMSENRKGYN NFRLNSGMPE YGQKGELRRD ERNL MWNID PYLQTQWQLS EKLSLDAGVR YSSVWFDSND HYVTPGNGDD SGDASYHKWL PAGSLKYAMT DAWNIYLAAG RGFET PTIN ELSYRADGQS GMNLGLKPST NDTIEIGSKT RIGDGLLSLA LFQTDTDDEI VVDSSSGGRT TYKNAGKTRR QGAELA WDQ RFAGDFRVNA SWTWLDATYR SNVCNEQDCN GNRMPGIARN MGFASIGYVP EDGWYAGTEA RYMGDIMADD ENTAKAP SY TLVGLFTGYK YNYHNLTVDL FGRVDNLFDK EYVGSVIVNE SNGRYYEPSP GRNYGVGMNI AWRFE UniProtKB: Pyrroloquinoline quinone transporter |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: PYRROLOQUINOLINE QUINONE
| Macromolecule | Name: PYRROLOQUINOLINE QUINONE / type: ligand / ID: 3 / Number of copies: 1 / Formula: PQQ |
|---|---|
| Molecular weight | Theoretical: 330.206 Da |
| Chemical component information | 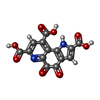 ChemComp-PQQ: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 111 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: LMNG was partly removed by concentration in a 100 kDa cut-off concentrator. | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 30 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 3 microliters of sample. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Number real images: 8744 / Average exposure time: 5.22 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)