[English] 日本語
 Yorodumi
Yorodumi- EMDB-45089: Carbon monoxide dehydrogenase (CODH) from Methanosarcina thermoph... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Carbon monoxide dehydrogenase (CODH) from Methanosarcina thermophila, specimen prepared on blot plunger | |||||||||
 Map data Map data | Carbon monoxide dehydrogenase (CODH) from Methanosarcina thermophila, specimen prepared on blot plunger | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CO-dehydrogenase / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethanogenesis, from acetate / anaerobic carbon monoxide dehydrogenase / anaerobic carbon-monoxide dehydrogenase activity / hydroxylamine reductase activity / acetyl-CoA metabolic process / nickel cation binding / peroxidase activity / response to hydrogen peroxide / 4 iron, 4 sulfur cluster binding / iron ion binding Similarity search - Function | |||||||||
| Biological species |   Methanosarcina thermophila (archaea) Methanosarcina thermophila (archaea) | |||||||||
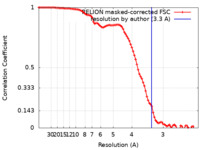
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Biester A / Drennan CL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Capturing a methanogenic carbon monoxide dehydrogenase/acetyl-CoA synthase complex via cryogenic electron microscopy. Authors: Alison Biester / David A Grahame / Catherine L Drennan /  Abstract: Approximately two-thirds of the estimated one-billion metric tons of methane produced annually by methanogens is derived from the cleavage of acetate. Acetate is broken down by a Ni-Fe-S-containing A- ...Approximately two-thirds of the estimated one-billion metric tons of methane produced annually by methanogens is derived from the cleavage of acetate. Acetate is broken down by a Ni-Fe-S-containing A-cluster within the enzyme acetyl-CoA synthase (ACS) to carbon monoxide (CO) and a methyl group (CH). The methyl group ultimately forms the greenhouse gas methane, whereas CO is converted to the greenhouse gas carbon dioxide (CO) by a Ni-Fe-S-containing C-cluster within the enzyme carbon monoxide dehydrogenase (CODH). Although structures have been solved of CODH/ACS from acetogens, which use these enzymes to make acetate from CO, no structure of a CODH/ACS from a methanogen has been reported. In this work, we use cryo-electron microscopy to reveal the structure of a methanogenic CODH and CODH/ACS from (CODH/ACS). We find that the N-terminal domain of acetogenic ACS, which is missing in all methanogens, is replaced by a domain of CODH. This CODH domain provides a channel for CO to travel between the two catalytic Ni-Fe-S clusters. It generates the binding surface for ACS and creates a remarkably similar CO alcove above the A-cluster using residues from CODH rather than ACS. Comparison of our CODH/ACS structure with our CODH structure reveals a molecular mechanism to restrict gas flow from the CO channel when ACS departs, preventing CO escape into the cell. Overall, these long-awaited structures of a methanogenic CODH/ACS reveal striking functional similarities to their acetogenic counterparts despite a substantial difference in domain organization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45089.map.gz emd_45089.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45089-v30.xml emd-45089-v30.xml emd-45089.xml emd-45089.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45089_fsc.xml emd_45089_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_45089.png emd_45089.png | 67.4 KB | ||
| Masks |  emd_45089_msk_1.map emd_45089_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45089.cif.gz emd-45089.cif.gz | 6.6 KB | ||
| Others |  emd_45089_half_map_1.map.gz emd_45089_half_map_1.map.gz emd_45089_half_map_2.map.gz emd_45089_half_map_2.map.gz | 48.6 MB 48.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45089 http://ftp.pdbj.org/pub/emdb/structures/EMD-45089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45089 | HTTPS FTP |
-Related structure data
| Related structure data |  9c0qMC  9c0rC  9c0sC  9c0tC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45089.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45089.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Carbon monoxide dehydrogenase (CODH) from Methanosarcina thermophila, specimen prepared on blot plunger | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45089_msk_1.map emd_45089_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_45089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_45089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Acetyl-CoA decarbonylase/synthase complex
| Entire | Name: Acetyl-CoA decarbonylase/synthase complex |
|---|---|
| Components |
|
-Supramolecule #1: Acetyl-CoA decarbonylase/synthase complex
| Supramolecule | Name: Acetyl-CoA decarbonylase/synthase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Carbon monoxide dehydrogenase heterotetramer, alpha and epsilon subunits |
|---|---|
| Source (natural) | Organism:   Methanosarcina thermophila (archaea) Methanosarcina thermophila (archaea) |
| Molecular weight | Theoretical: 212 KDa |
-Macromolecule #1: Acetyl-CoA decarbonylase/synthase complex subunit alpha 2
| Macromolecule | Name: Acetyl-CoA decarbonylase/synthase complex subunit alpha 2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: anaerobic carbon monoxide dehydrogenase |
|---|---|
| Source (natural) | Organism:   Methanosarcina thermophila (archaea) Methanosarcina thermophila (archaea) |
| Molecular weight | Theoretical: 87.855852 KDa |
| Sequence | String: MSKLTTGSFS IEDLESVQIT INNIVGAAKE AAEKAEEELG PMGPTPFPTA ATVRDWSFTL FDRYEPVYTP MCDQCCYCTF GPCNLEGNR RGACGLDMKG QAAREFFLRC ITGCACHSAH GRHLLDHIIS IFGEDMPINM GASNVIAPNI QLITGRQPKT L GDLKPIME ...String: MSKLTTGSFS IEDLESVQIT INNIVGAAKE AAEKAEEELG PMGPTPFPTA ATVRDWSFTL FDRYEPVYTP MCDQCCYCTF GPCNLEGNR RGACGLDMKG QAAREFFLRC ITGCACHSAH GRHLLDHIIS IFGEDMPINM GASNVIAPNI QLITGRQPKT L GDLKPIME YVEEELGQLL ATVHAGQEGA AIDYDNKAML AGILDHVGME VSDIAQVTAL GFPKSDPEAP LVEVGMGTLD AS KPVIIAI GHNVAGVTYI MDYMEDNNLT DKMEIGGLCC TAFDMTRYKR EDRKPPYAKI VGTISKELKV VRSGIPDVIV IDE QCVRAD LVEEGKKLKI PVIASNEKVM YGLPDRTNDD VDAIIEDIKT GKIPGCVMLD YEKLGELVPR LAMEMAPLRE GISA IPSDE EMASLVAKCV ACGECALACP EELDIPDAIQ AAKEGDFTAL DFLHDLCVGC RRCEQVCNKE IPILSVIDKA AQKAI AEEK GLVRAGRGQV SDAEIRAEGL NLVMGTTPGV IAIIGCANYP AGSKDVYRIA EEFLNRNYIV AVSGCSAMDI GMYKDA DGK TLYERFPGRF ERGNILNTGS CVSNSHISGT CHKVAAIFAG RNLSGNLAEI ADYTLNRVGA VGLAWGAYSQ KAAAIGT GC NMYGIPAVLG PHSGKYRRAL IAKTYDENKW KVYDSRNGSE LDIPPSPEFL ITTAETWQEA CVLLAKNCIR PSDNNMGR S IKLTHWIELS EKYLGVLPED WWKFVRHEAD LPLSRREELL KKLETEHGWE IDWKKKKIIS GPKIKFDVSS QPTNLKRLC KEA UniProtKB: Acetyl-CoA decarbonylase/synthase complex subunit alpha 2 |
-Macromolecule #2: Acetyl-CoA decarbonylase/synthase complex subunit epsilon 2
| Macromolecule | Name: Acetyl-CoA decarbonylase/synthase complex subunit epsilon 2 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Methanosarcina thermophila (archaea) Methanosarcina thermophila (archaea) |
| Molecular weight | Theoretical: 18.587375 KDa |
| Sequence | String: MVDTTKNTKL FTSYGVKTSK AITTEVAAKL ISKAKRPLFV VGTGVLDPEL LDRAVKIAKA KNIPIAATGS SMPGFVDKDV NAKYINLHQ LGFYLTDPDW PGLDGNGNYD TIILLGHKKY YINQVLSAVK NFSDVKSISI DRNYIQNATM SFGNLSKADH I AALDEVID LL UniProtKB: Acetyl-CoA decarbonylase/synthase complex subunit epsilon 2 |
-Macromolecule #3: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 3 / Number of copies: 7 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #4: Fe(3)-Ni(1)-S(4) cluster
| Macromolecule | Name: Fe(3)-Ni(1)-S(4) cluster / type: ligand / ID: 4 / Number of copies: 2 / Formula: RQM |
|---|---|
| Molecular weight | Theoretical: 410.333 Da |
| Chemical component information |  ChemComp-RQM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.53 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Software | Name:  Coot (ver. 0.9.8.92) Coot (ver. 0.9.8.92) |
| Details | Initial model was generated using AlphaFold, fit to the map using ChimeraX, followed by rigid body fitting in phenix, and finally iterative real space refinement in coot and phenix. |
| Refinement | Space: REAL / Overall B value: 90.5204 / Target criteria: Correlation coefficient |
| Output model |  PDB-9c0q: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)