[English] 日本語
 Yorodumi
Yorodumi- EMDB-44644: SARS CoV2 spike in complex with NTD-directed antibody Fab DH1052 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS CoV2 spike in complex with NTD-directed antibody Fab DH1052 | |||||||||
 Map data Map data | CoV2 spike in complex with antibody Fab DH1052 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS coronavirus spike antibody / VIRAL PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
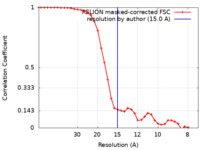
| Method | single particle reconstruction / negative staining / Resolution: 15.0 Å | |||||||||
 Authors Authors | Edwards RJ / Mansouri K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Non-neutralizing SARS-CoV-2 N-terminal domain antibodies protect mice against severe disease using Fc-mediated effector functions. Authors: Camille N Pierre / Lily E Adams / Jaclyn S Higgins / Kara Anasti / Derrick Goodman / Dieter Mielke / Sherry Stanfield-Oakley / John M Powers / Dapeng Li / Wes Rountree / Yunfei Wang / Robert ...Authors: Camille N Pierre / Lily E Adams / Jaclyn S Higgins / Kara Anasti / Derrick Goodman / Dieter Mielke / Sherry Stanfield-Oakley / John M Powers / Dapeng Li / Wes Rountree / Yunfei Wang / Robert J Edwards / S Munir Alam / Guido Ferrari / Georgia D Tomaras / Barton F Haynes / Ralph S Baric / Kevin O Saunders /  Abstract: Antibodies perform both neutralizing and non-neutralizing effector functions that protect against certain pathogen-induced diseases. A human antibody directed at the SARS-CoV-2 Spike N-terminal ...Antibodies perform both neutralizing and non-neutralizing effector functions that protect against certain pathogen-induced diseases. A human antibody directed at the SARS-CoV-2 Spike N-terminal domain (NTD), DH1052, was recently shown to be non-neutralizing, yet it protected mice and cynomolgus macaques from severe disease. The mechanisms of NTD non-neutralizing antibody-mediated protection are unknown. Here we show that Fc effector functions mediate NTD non-neutralizing antibody (non-nAb) protection against SARS-CoV-2 MA10 viral challenge in mice. Though non-nAb prophylactic infusion did not suppress infectious viral titers in the lung as potently as neutralizing antibody (nAb) infusion, disease markers including gross lung discoloration were similar in nAb and non-nAb groups. Fc functional knockout substitutions abolished non-nAb protection and increased viral titers in the nAb group. Fc enhancement increased non-nAb protection relative to WT, supporting a positive association between Fc functionality and degree of protection from SARS-CoV-2 infection. For therapeutic administration of antibodies, non-nAb effector functions contributed to virus suppression and lessening of lung discoloration, but the presence of neutralization was required for optimal protection from disease. This study demonstrates that non-nAbs can utilize Fc-mediated mechanisms to lower viral load and prevent lung damage due to coronavirus infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44644.map.gz emd_44644.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44644-v30.xml emd-44644-v30.xml emd-44644.xml emd-44644.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44644_fsc.xml emd_44644_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_44644.png emd_44644.png | 51.6 KB | ||
| Filedesc metadata |  emd-44644.cif.gz emd-44644.cif.gz | 5.2 KB | ||
| Others |  emd_44644_half_map_1.map.gz emd_44644_half_map_1.map.gz emd_44644_half_map_2.map.gz emd_44644_half_map_2.map.gz | 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44644 http://ftp.pdbj.org/pub/emdb/structures/EMD-44644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44644 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44644.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44644.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CoV2 spike in complex with antibody Fab DH1052 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_44644_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
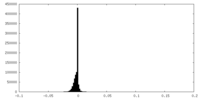
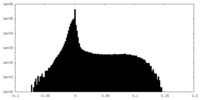
| Density Histograms |
-Half map: Half map 2
| File | emd_44644_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
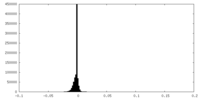
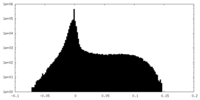
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of SARS CoV2 spike with NTD-directed antibody Fab DH1052
| Entire | Name: Complex of SARS CoV2 spike with NTD-directed antibody Fab DH1052 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of SARS CoV2 spike with NTD-directed antibody Fab DH1052
| Supramolecule | Name: Complex of SARS CoV2 spike with NTD-directed antibody Fab DH1052 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 372 KDa |
-Supramolecule #2: DH1052 antibody Fab
| Supramolecule | Name: DH1052 antibody Fab / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: SARS CoV2 spike
| Supramolecule | Name: SARS CoV2 spike / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl formate Details: A 5 microliter drop of complex was incubated on the grid for 10 to 15 s, blotted and then stained with 2 percent uranyl formate for 1 minute, then blotted and air dried. | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | |||||||||
| Details | Spike protein and Fab protein at 1 mg per ml were stored at minus 80 degrees C until use. Spike was thawed in an aluminum block at 37 degrees C for 5 minutes, Fab was thawed in an aluminum block at room temperature for 5 minutes. To for the complex, 2 microliters of spike was mixed with Fab at a 1 to 9 ratio spike to Fab, and incubated at 37 degrees C for 1 hour. The complex was then crosslinked by diluting to a final spike concentration of 0.1 mg per ml into room temperature buffer containing 150 mM NaCl, 20 mM HEPES pH 7.4, 5 percent glycerol, and 7.5 mM glutaraldehyde. After 5 minutes cross-linking, excess glutaraldehyde was quenched by adding sufficient 1 M Tris pH 7.4 stock to give a final concentration of 75 mM Tris and incubated for 5 minutes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 200 / Average exposure time: 0.5 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 82000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)