[English] 日本語
 Yorodumi
Yorodumi- EMDB-44357: Human polymerase epsilon bound to PCNA and DNA in the nucleotide ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human polymerase epsilon bound to PCNA and DNA in the nucleotide exchange state | |||||||||
 Map data Map data | Final EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA polymerase / DNA / DNA Binding Protein-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA replication initiation / epsilon DNA polymerase complex / positive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication / purine-specific mismatch base pair DNA N-glycosylase activity / nucleotide-excision repair, DNA gap filling / nuclear lamina / Polymerase switching ...DNA replication initiation / epsilon DNA polymerase complex / positive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication / purine-specific mismatch base pair DNA N-glycosylase activity / nucleotide-excision repair, DNA gap filling / nuclear lamina / Polymerase switching / positive regulation of DNA-directed DNA polymerase activity / Processive synthesis on the lagging strand / DNA replication proofreading / MutLalpha complex binding / PCNA complex / single-stranded DNA 3'-5' DNA exonuclease activity / Telomere C-strand (Lagging Strand) Synthesis / Removal of the Flap Intermediate / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Transcription of E2F targets under negative control by DREAM complex / Polymerase switching on the C-strand of the telomere / replisome / Processive synthesis on the C-strand of the telomere / response to L-glutamate / Removal of the Flap Intermediate from the C-strand / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / response to dexamethasone / DNA synthesis involved in DNA repair / histone acetyltransferase binding / DNA polymerase processivity factor activity / leading strand elongation / G1/S-Specific Transcription / nuclear replication fork / replication fork processing / SUMOylation of DNA replication proteins / PCNA-Dependent Long Patch Base Excision Repair / Activation of the pre-replicative complex / embryonic organ development / response to cadmium ion / estrous cycle / mismatch repair / translesion synthesis / cyclin-dependent protein kinase holoenzyme complex / base-excision repair, gap-filling / DNA polymerase binding / epithelial cell differentiation / liver regeneration / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / positive regulation of DNA replication / nuclear estrogen receptor binding / replication fork / positive regulation of DNA repair / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / male germ cell nucleus / Termination of translesion DNA synthesis / G1/S transition of mitotic cell cycle / Translesion Synthesis by POLH / Recognition of DNA damage by PCNA-containing replication complex / receptor tyrosine kinase binding / HDR through Homologous Recombination (HRR) / DNA-templated DNA replication / cellular response to xenobiotic stimulus / Dual Incision in GG-NER / cellular response to hydrogen peroxide / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / response to estradiol / mitotic cell cycle / E3 ubiquitin ligases ubiquitinate target proteins / heart development / 4 iron, 4 sulfur cluster binding / chromatin organization / DNA-directed DNA polymerase / damaged DNA binding / DNA-directed DNA polymerase activity / DNA replication / chromosome, telomeric region / nuclear body / nucleotide binding / chromatin binding / centrosome / chromatin / protein-containing complex binding / enzyme binding / negative regulation of transcription by RNA polymerase II / DNA binding / extracellular exosome / zinc ion binding / nucleoplasm / identical protein binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / DNA molecule (others) Homo sapiens (human) / DNA molecule (others) | |||||||||
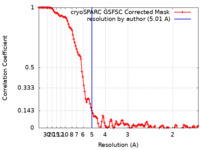
| Method | single particle reconstruction / cryo EM / Resolution: 5.01 Å | |||||||||
 Authors Authors | Wang F / He Q / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structures of the human leading strand Polε-PCNA holoenzyme. Authors: Qing He / Feng Wang / Nina Y Yao / Michael E O'Donnell / Huilin Li /  Abstract: In eukaryotes, the leading strand DNA is synthesized by Polε and the lagging strand by Polδ. These replicative polymerases have higher processivity when paired with the DNA clamp PCNA. While the ...In eukaryotes, the leading strand DNA is synthesized by Polε and the lagging strand by Polδ. These replicative polymerases have higher processivity when paired with the DNA clamp PCNA. While the structure of the yeast Polε catalytic domain has been determined, how Polε interacts with PCNA is unknown in any eukaryote, human or yeast. Here we report two cryo-EM structures of human Polε-PCNA-DNA complex, one in an incoming nucleotide bound state and the other in a nucleotide exchange state. The structures reveal an unexpected three-point interface between the Polε catalytic domain and PCNA, with the conserved PIP (PCNA interacting peptide)-motif, the unique P-domain, and the thumb domain each interacting with a different protomer of the PCNA trimer. We propose that the multi-point interface prevents other PIP-containing factors from recruiting to PCNA while PCNA functions with Polε. Comparison of the two states reveals that the finger domain pivots around the [4Fe-4S] cluster-containing tip of the P-domain to regulate nucleotide exchange and incoming nucleotide binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44357.map.gz emd_44357.map.gz | 90.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44357-v30.xml emd-44357-v30.xml emd-44357.xml emd-44357.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44357_fsc.xml emd_44357_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_44357.png emd_44357.png | 52 KB | ||
| Filedesc metadata |  emd-44357.cif.gz emd-44357.cif.gz | 8 KB | ||
| Others |  emd_44357_additional_1.map.gz emd_44357_additional_1.map.gz emd_44357_half_map_1.map.gz emd_44357_half_map_1.map.gz emd_44357_half_map_2.map.gz emd_44357_half_map_2.map.gz | 87.8 MB 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44357 http://ftp.pdbj.org/pub/emdb/structures/EMD-44357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44357 | HTTPS FTP |
-Related structure data
| Related structure data |  9b8sMC  9b8tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44357.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44357.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
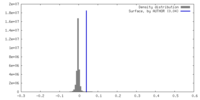
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened EM map
| File | emd_44357_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
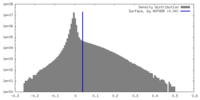
| Density Histograms |
-Half map: Half Map A
| File | emd_44357_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
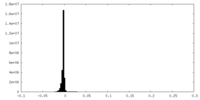
| Density Histograms |
-Half map: Half map B
| File | emd_44357_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
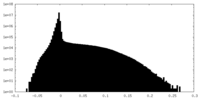
| Density Histograms |
- Sample components
Sample components
-Entire : The DNA bound Pol epsilon and PCNA complex
| Entire | Name: The DNA bound Pol epsilon and PCNA complex |
|---|---|
| Components |
|
-Supramolecule #1: The DNA bound Pol epsilon and PCNA complex
| Supramolecule | Name: The DNA bound Pol epsilon and PCNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: DNA polymerase epsilon catalytic subunit A
| Macromolecule | Name: DNA polymerase epsilon catalytic subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 261.753234 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MSLRSGGRRR ADPGADGEAS RDDGATSSVS ALKRLERSQW TDKMDLRFGF ERLKEPGEKT GWLINMHPTE ILDEDKRLGS AVDYYFIQD DGSRFKVALP YKPYFYIATR KGCEREVSSF LSKKFQGKIA KVETVPKEDL DLPNHLVGLK RNYIRLSFHT V EDLVKVRK ...String: MSLRSGGRRR ADPGADGEAS RDDGATSSVS ALKRLERSQW TDKMDLRFGF ERLKEPGEKT GWLINMHPTE ILDEDKRLGS AVDYYFIQD DGSRFKVALP YKPYFYIATR KGCEREVSSF LSKKFQGKIA KVETVPKEDL DLPNHLVGLK RNYIRLSFHT V EDLVKVRK EISPAVKKNR EQDHASDAYT ALLSSVLQRG GVITDEEETS KKIADQLDNI VDMREYDVPY HIRLSIDLKI HV AHWYNVR YRGNAFPVEI TRRDDLVERP DPVVLAFAIA TTKLPLKFPD AETDQIMMIS YMIDGQGYLI TNREIVSEDI EDF EFTPKP EYEGPFCVFN EPDEAHLIQR WFEHVQETKP TIMVTYNGDF FDWPFVEARA AVHGLSMQQE IGFQKDSQGE YKAP QCIHM DCLRWVKRDS YLPVGSHNLK AAAKAKLGYD PVELDPEDMC RMATEQPQTL ATYSVSDAVA TYYLYMKYVH PFIFA LCTI IPMEPDEVLR KGSGTLCEAL LMVQAFHANI IFPNKQEQEF NKLTDDGHVL DSETYVGGHV EALESGVFRS DIPCRF RMN PAAFDFLLQR VEKTLRHALE EEEKVPVEQV TNFEEVCDEI KSKLASLKDV PSRIECPLIY HLDVGAMYPN IILTNRL QP SAMVDEATCA ACDFNKPGAN CQRKMAWQWR GEFMPASRSE YHRIQHQLES EKFPPLFPEG PARAFHELSR EEQAKYEK R RLADYCRKAY KKIHITKVEE RLTTICQREN SFYVDTVRAF RDRRYEFKGL HKVWKKKLSA AVEVGDAAEV KRCKNMEVL YDSLQLAHKC ILNSFYGYVM RKGARWYSME MAGIVCFTGA NIITQARELI EQIGRPLELD TDGIWCVLPN SFPENFVFKT TNVKKPKVT ISYPGAMLNI MVKEGFTNDQ YQELAEPSSL TYVTRSENSI FFEVDGPYLA MILPASKEEG KKLKKRYAVF N EDGSLAEL KGFEVKRRGE LQLIKIFQSS VFEAFLKGST LEEVYGSVAK VADYWLDVLY SKAANMPDSE LFELISENRS MS RKLEDYG EQKSTSISTA KRLAEFLGDQ MVKDAGLSCR YIISRKPEGS PVTERAIPLA IFQAEPTVRK HFLRKWLKSS SLQ DFDIRA ILDWDYYIER LGSAIQKIIT IPAALQQVKN PVPRVKHPDW LHKKLLEKND VYKQKKISEL FTLEGRRQVT MAEA SEDSP RPSAPDMEDF GLVKLPHPAA PVTVKRKRVL WESQEESQDL TPTVPWQEIL GQPPALGTSQ EEWLVWLRFH KKKWQ LQAR QRLARRKRQR LESAEGVLRP GAIRDGPATG LGSFLRRTAR SILDLPWQIV QISETSQAGL FRLWALVGSD LHCIRL SIP RVFYVNQRVA KAEEGASYRK VNRVLPRSNM VYNLYEYSVP EDMYQEHINE INAELSAPDI EGVYETQVPL LFRALVH LG CVCVVNKQLV RHLSGWEAET FALEHLEMRS LAQFSYLEPG SIRHIYLYHH AQAHKALFGI FIPSQRRASV FVLDTVRS N QMPSLGALYS AEHGLLLEKV GPELLPPPKH TFEVRAETDL KTICRAIQRF LLAYKEERRG PTLIAVQSSW ELKRLASEI PVLEEFPLVP ICVADKINYG VLDWQRHGAR RMIRHYLNLD TCLSQAFEMS RYFHIPIGNL PEDISTFGSD LFFARHLQRH NHLLWLSPT ARPDLGGKEA DDNCLVMEFD DQATVEINSS GCYSTVCVEL DLQNLAVNTI LQSHHVNDME GADSMGISFD V IQQASLED MITGGQAASA PASYDETALC SNTFRILKSM VVGWVKEITQ YHNIYADNQV MHFYRWLRSP SSLLHDPALH RT LHNMMKK LFLQLIAEFK RLGSSVIYAN FNRIILCTKK RRVEDAIAYV EYITSSIHSK ETFHSLTISF SRCWEFLLWM DPS NYGGIK GKVSSRIHCG LQDSQKAGGA EDEQENEDDE EERDGEEEEE AEESNVEDLL ENNWNILQFL PQAASCQNYF LMIV SAYIV AVYHCMKDGL RRSAPGSTPV RRRGASQLSQ EAEGAVGALP GMITFSQDYV ANELTQSFFT ITQKIQKKVT GSRNS TELS EMFPVLPGSH LLLNNPALEF IKYVCKVLSL DTNITNQVNK LNRDLLRLVD VGEFSEEAQF RDPCRSYVLP EVICRS CNF CRDLDLCKDS SFSEDGAVLP QWLCSNCQAP YDSSAIEMTL VEVLQKKLMA FTLQDLVCLK CRGVKETSMP VYCSCAG DF ALTIHTQVFM EQIGIFRNIA QHYGMSYLLE TLEWLLQKNP QLGH UniProtKB: DNA polymerase epsilon catalytic subunit A |
-Macromolecule #2: Proliferating cell nuclear antigen
| Macromolecule | Name: Proliferating cell nuclear antigen / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.795752 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MFEARLVQGS ILKKVLEALK DLINEACWDI SSSGVNLQSM DSSHVSLVQL TLRSEGFDTY RCDRNLAMGV NLTSMSKILK CAGNEDIIT LRAEDNADTL ALVFEAPNQE KVSDYEMKLM DLDVEQLGIP EQEYSCVVKM PSGEFARICR DLSHIGDAVV I SCAKDGVK ...String: MFEARLVQGS ILKKVLEALK DLINEACWDI SSSGVNLQSM DSSHVSLVQL TLRSEGFDTY RCDRNLAMGV NLTSMSKILK CAGNEDIIT LRAEDNADTL ALVFEAPNQE KVSDYEMKLM DLDVEQLGIP EQEYSCVVKM PSGEFARICR DLSHIGDAVV I SCAKDGVK FSASGELGNG NIKLSQTSNV DKEEEAVTIE MNEPVQLTFA LRYLNFFTKA TPLSSTVTLS MSADVPLVVE YK IADMGHL KYYLAPKIED EEGS UniProtKB: Proliferating cell nuclear antigen |
-Macromolecule #3: DNA (5'-D(P*GP*TP*GP*AP*TP*GP*CP*TP*TP*TP*AP*GP*AP*TP*TP*TP*TP*TP...
| Macromolecule | Name: DNA (5'-D(P*GP*TP*GP*AP*TP*GP*CP*TP*TP*TP*AP*GP*AP*TP*TP*TP*TP*TP*C)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 10.823965 KDa |
| Sequence | String: (DT)(DG)(DA)(DG)(DG)(DT)(DT)(DC)(DA)(DG) (DC)(DA)(DA)(DG)(DG)(DT)(DG)(DA)(DT)(DG) (DC)(DT)(DT)(DT)(DA)(DG)(DA)(DT)(DT) (DT)(DT)(DT)(DC)(DA)(DC) |
-Macromolecule #4: DNA (5'-D(P*AP*AP*AP*GP*TP*GP*AP*AP*AP*AP*AP*TP*CP*TP*AP*AP*AP*GP...
| Macromolecule | Name: DNA (5'-D(P*AP*AP*AP*GP*TP*GP*AP*AP*AP*AP*AP*TP*CP*TP*AP*AP*AP*GP*CP*AP*TP*CP*AP*C)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 18.13668 KDa |
| Sequence | String: (DG)(DC)(DC)(DA)(DC)(DG)(DC)(DT)(DG)(DA) (DG)(DA)(DG)(DC)(DC)(DA)(DG)(DC)(DA)(DG) (DC)(DA)(DA)(DA)(DG)(DT)(DG)(DA)(DA) (DA)(DA)(DA)(DT)(DC)(DT)(DA)(DA)(DA)(DG) (DC) (DA)(DT)(DC)(DA)(DC)(DC) ...String: (DG)(DC)(DC)(DA)(DC)(DG)(DC)(DT)(DG)(DA) (DG)(DA)(DG)(DC)(DC)(DA)(DG)(DC)(DA)(DG) (DC)(DA)(DA)(DA)(DG)(DT)(DG)(DA)(DA) (DA)(DA)(DA)(DT)(DC)(DT)(DA)(DA)(DA)(DG) (DC) (DA)(DT)(DC)(DA)(DC)(DC)(DT)(DT) (DG)(DC)(DT)(DG)(DA)(DA)(DC)(DC)(DT)(DC) (DA) |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 280 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-9b8s: |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)