+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rat TRPV2 E724A/D725A bound to 2-APB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPV2 / TRP channel / ion channel / 2-APB / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgrowth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / calcium ion import across plasma membrane / axonal growth cone / positive regulation of axon extension / monoatomic cation channel activity / endomembrane system / calcium channel activity ...growth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / calcium ion import across plasma membrane / axonal growth cone / positive regulation of axon extension / monoatomic cation channel activity / endomembrane system / calcium channel activity / melanosome / lamellipodium / positive regulation of cold-induced thermogenesis / cell body / axon / negative regulation of cell population proliferation / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Pumroy RA / Rocereta JA / Moiseenkova-Bell VY | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural insights into TRPV2 modulation by probenecid. Authors: Julia A Rocereta / Toni Sturhahn / Ruth A Pumroy / Tabea C Fricke / Christine Herzog / Andreas Leffler / Vera Moiseenkova-Bell /   Abstract: The transient receptor potential vanilloid 2 (TRPV2) cation channel is a key player in cardiovascular physiology and pathophysiology. Probenecid (PBC), an FDA-approved uricosuric agent thought to ...The transient receptor potential vanilloid 2 (TRPV2) cation channel is a key player in cardiovascular physiology and pathophysiology. Probenecid (PBC), an FDA-approved uricosuric agent thought to activate TRPV2, has shown promise in enhancing cardiovascular function in both preclinical and clinical studies. Here our electrophysiological data reveal that PBC significantly potentiates rat TRPV2 to known stimuli, and cryo electron microscopy structures show that PBC directly interacts with rat TRPV2 in a previously unidentified intracellular binding pocket. PBC binding at a conserved TRPV2-specific histidine prevents the channel from taking on the inactivated carboxyl-terminal conformation. This effect extends to TRPV1 and TRPV3 channels when glutamine is substituted with histidine at the corresponding position, increasing their sensitivity to PBC. While PBC alone does not induce TRPV2 opening, its combination with 2-aminoethoxydiphenyl borate enables the channel to adopt an intermediate, potentiated state. Our results offer insights into potential therapeutic advancements for TRPV2 through this pocket. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44159.map.gz emd_44159.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44159-v30.xml emd-44159-v30.xml emd-44159.xml emd-44159.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44159.png emd_44159.png | 115.1 KB | ||
| Filedesc metadata |  emd-44159.cif.gz emd-44159.cif.gz | 6.3 KB | ||
| Others |  emd_44159_half_map_1.map.gz emd_44159_half_map_1.map.gz emd_44159_half_map_2.map.gz emd_44159_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44159 http://ftp.pdbj.org/pub/emdb/structures/EMD-44159 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44159 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44159 | HTTPS FTP |
-Validation report
| Summary document |  emd_44159_validation.pdf.gz emd_44159_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44159_full_validation.pdf.gz emd_44159_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_44159_validation.xml.gz emd_44159_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_44159_validation.cif.gz emd_44159_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44159 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44159 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44159 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44159 | HTTPS FTP |
-Related structure data
| Related structure data |  9b3xMC  9b3uC  9b3vC  9b3wC  9b3yC  9b3zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44159.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44159.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_44159_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_44159_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetramer of rat TRPV2 E724A/D725A mutant
| Entire | Name: Homotetramer of rat TRPV2 E724A/D725A mutant |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer of rat TRPV2 E724A/D725A mutant
| Supramolecule | Name: Homotetramer of rat TRPV2 E724A/D725A mutant / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 2
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 2 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.696844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSASSPPAF RLETSDGDEE GNAEVNKGKQ EPPPMESPFQ REDRNSSPQI KVNLNFIKRP PKNTSAPSQQ EPDRFDRDRL FSVVSRGVP EELTGLLEYL RWNSKYLTDS AYTEGSTGKT CLMKAVLNLQ DGVNACIMPL LQIDKDSGNP KLLVNAQCTD E FYQGHSAL ...String: MTSASSPPAF RLETSDGDEE GNAEVNKGKQ EPPPMESPFQ REDRNSSPQI KVNLNFIKRP PKNTSAPSQQ EPDRFDRDRL FSVVSRGVP EELTGLLEYL RWNSKYLTDS AYTEGSTGKT CLMKAVLNLQ DGVNACIMPL LQIDKDSGNP KLLVNAQCTD E FYQGHSAL HIAIEKRSLQ CVKLLVENGA DVHLRACGRF FQKHQGTCFY FGELPLSLAA CTKQWDVVTY LLENPHQPAS LE ATDSLGN TVLHALVMIA DNSPENSALV IHMYDGLLQM GARLCPTVQL EEISNHQGLT PLKLAAKEGK IEIFRHILQR EFS GPYQPL SRKFTEWCYG PVRVSLYDLS SVDSWEKNSV LEIIAFHCKS PNRHRMVVLE PLNKLLQEKW DRLVSRFFFN FACY LVYMF IFTVVAYHQP SLDQPAIPSS KATFGESMLL LGHILILLGG IYLLLGQLWY FWRRRLFIWI SFMDSYFEIL FLLQA LLTV LSQVLRFMET EWYLPLLVLS LVLGWLNLLY YTRGFQHTGI YSVMIQKVIL RDLLRFLLVY LVFLFGFAVA LVSLSR EAR SPKAPEDNNS TVTEQPTVGQ EEEPAPYRSI LDASLELFKF TIGMGELAFQ EQLRFRGVVL LLLLAYVLLT YVLLLNM LI ALMSETVNHV ADNSWSIWKL QKAISVLEME NGYWWCRRKK HREGRLLKVG TRGDGTPDER WCFRVEEVNW AAWEKTLP T LSAAPSGPGI TGNKKNPTSK PGKNSASEED HLPLQVLQSP UniProtKB: Transient receptor potential cation channel subfamily V member 2 |
-Macromolecule #2: 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE
| Macromolecule | Name: 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE / type: ligand / ID: 2 / Number of copies: 4 / Formula: PEX |
|---|---|
| Molecular weight | Theoretical: 522.632 Da |
| Chemical component information | 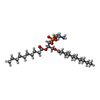 ChemComp-PEX: |
-Macromolecule #3: 2-aminoethyl diphenylborinate
| Macromolecule | Name: 2-aminoethyl diphenylborinate / type: ligand / ID: 3 / Number of copies: 4 / Formula: FZ4 |
|---|---|
| Molecular weight | Theoretical: 225.094 Da |
| Chemical component information | 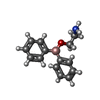 ChemComp-FZ4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































