[English] 日本語
 Yorodumi
Yorodumi- EMDB-44046: Cryo-EM structure of Acanthamoeba polyphaga mimivirus Fanzor2 ter... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
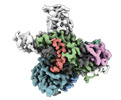
| Title | Cryo-EM structure of Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transposon / Fanzor / cryo-EM / DNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus | |||||||||
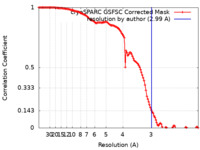
| Method | single particle reconstruction / cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Qayyum MZ / Schargel RD / Tanwar AS / Kellogg EH / Kalathur RC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structure of Fanzor2 reveals insights into the evolution of the TnpB superfamily. Authors: Richard D Schargel / M Zuhaib Qayyum / Ajay Singh Tanwar / Ravi C Kalathur / Elizabeth H Kellogg /  Abstract: RNA-guided endonucleases, once thought to be exclusive to prokaryotes, have been recently identified in eukaryotes and are called Fanzors. They are classified into two clades, Fanzor1 and Fanzor2. ...RNA-guided endonucleases, once thought to be exclusive to prokaryotes, have been recently identified in eukaryotes and are called Fanzors. They are classified into two clades, Fanzor1 and Fanzor2. Here we present the cryo-electron microscopy structure of Acanthamoeba polyphaga mimivirus Fanzor2, revealing its ωRNA architecture, active site and features involved in transposon-adjacent motif recognition. A comparison to Fanzor1 and TnpB structures highlights divergent evolutionary paths, advancing our understanding of RNA-guided endonucleases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44046.map.gz emd_44046.map.gz | 769.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44046-v30.xml emd-44046-v30.xml emd-44046.xml emd-44046.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44046_fsc.xml emd_44046_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_44046.png emd_44046.png | 110.4 KB | ||
| Filedesc metadata |  emd-44046.cif.gz emd-44046.cif.gz | 6.6 KB | ||
| Others |  emd_44046_half_map_1.map.gz emd_44046_half_map_1.map.gz emd_44046_half_map_2.map.gz emd_44046_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44046 http://ftp.pdbj.org/pub/emdb/structures/EMD-44046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44046 | HTTPS FTP |
-Related structure data
| Related structure data |  9b0lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44046.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44046.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.044 Å | ||||||||||||||||||||||||||||||||||||
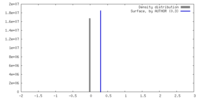
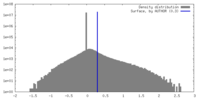
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_44046_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
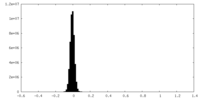
| Density Histograms |
-Half map: #1
| File | emd_44046_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
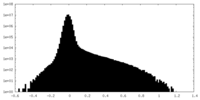
| Density Histograms |
- Sample components
Sample components
-Entire : Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex
| Entire | Name: Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex
| Supramolecule | Name: Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
-Macromolecule #1: DNA (5'-D(P*GP*AP*CP*GP*GP*TP*CP*GP*GP*GP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*GP*AP*CP*GP*GP*TP*CP*GP*GP*GP*C)-3') / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 3.415222 KDa |
| Sequence | String: (DG)(DA)(DC)(DG)(DG)(DT)(DC)(DG)(DG)(DG) (DC) |
-Macromolecule #3: DNA (5'-D(P*CP*CP*CP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*CP*CP*A)-3') / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 1.135795 KDa |
| Sequence | String: (DC)(DC)(DC)(DA) |
-Macromolecule #4: DNA (5'-D(P*AP*GP*GP*AP*GP*GP*TP*GP*GP*CP*TP*GP*CP*CP*CP*CP*GP*AP...
| Macromolecule | Name: DNA (5'-D(P*AP*GP*GP*AP*GP*GP*TP*GP*GP*CP*TP*GP*CP*CP*CP*CP*GP*AP*CP*CP*GP*TP*C)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 7.083547 KDa |
| Sequence | String: (DA)(DG)(DG)(DA)(DG)(DG)(DT)(DG)(DG)(DC) (DT)(DG)(DC)(DC)(DC)(DC)(DG)(DA)(DC)(DC) (DG)(DT)(DC) |
-Macromolecule #2: TnpB-like protein L79
| Macromolecule | Name: TnpB-like protein L79 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 60.572387 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKEAVKNVKP KVPAKKRIIT GSKTKKKVFV KKKPPDKKPL KKPVKKTVKT DKPKSIYVPN KDLKISKWIP TPKKEFTEIE TNSWYEHRK FENPNKSPVQ TYNKIVPVVP PESIKQQNLA NKRKKTNRPI VFISSEKIRI YPTKDQQKIL QTWFRLFAYM Y NCTIDYIN ...String: MKEAVKNVKP KVPAKKRIIT GSKTKKKVFV KKKPPDKKPL KKPVKKTVKT DKPKSIYVPN KDLKISKWIP TPKKEFTEIE TNSWYEHRK FENPNKSPVQ TYNKIVPVVP PESIKQQNLA NKRKKTNRPI VFISSEKIRI YPTKDQQKIL QTWFRLFAYM Y NCTIDYIN SKKVVLESGR INVAATRKVC NKISVRKAQK TIRDNLIQST NPSIMTHIID EAIGLACSNY KTCLTNYIER HI KKFDIKP WNMSKRKKII IIEANFFKKG TFCPTVFPKM ESSKPLTMID KTVTLQYDSD TRKYILFVPR VTPKYSVNKE KNS CGIDPG LRDFLTVYSE NETQSICPIE IVVNTTKNEY KKIDKINEII KTKPNLNSKR KKKLNRGLRK YHRRVTNKMK DMHY KVSHE LVNTFDKICI GKLNVKSILS KANTVLKSAL KRKLATLSFY RFTQRLTHMG YKYGTEVVNV NEYLTTKTCS NCGKI KDLG ASKIYECESC GMYADRDENA AKNILKVGLK PWYKQK UniProtKB: TnpB-like protein L79 |
-Macromolecule #5: RNA (117-MER)
| Macromolecule | Name: RNA (117-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 79.330805 KDa |
| Sequence | String: AAAAAUAGUC UAAUAAAAUC AGGGGUACAU UCCGCUAGUA CUCCACCCUA CGGGUUAAGC AAAUGAGAAU AUCGAAACGG UAUGCACAG GAUUCUUCGA GUGAUAAUCU UAGGAUGACU CACUAAGGAG AUGACUAAAG UGUAUCAUUC AAUAUUGUAU U GAACGGUA ...String: AAAAAUAGUC UAAUAAAAUC AGGGGUACAU UCCGCUAGUA CUCCACCCUA CGGGUUAAGC AAAUGAGAAU AUCGAAACGG UAUGCACAG GAUUCUUCGA GUGAUAAUCU UAGGAUGACU CACUAAGGAG AUGACUAAAG UGUAUCAUUC AAUAUUGUAU U GAACGGUA UUCUUCCAUA GAGAGUUGAU UUUUGGAGUA UCCAAAAAUA UCAACUUUUU AUGAGCGGGC AGCCACCUCC UU GUUAUUG GENBANK: GENBANK: AY653733.1 |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)