[English] 日本語
 Yorodumi
Yorodumi- EMDB-43797: Local refinement of 5-HT2AR bound to 5-HT in complex with a mini-... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of 5-HT2AR bound to 5-HT in complex with a mini-Gq protein and scFv16 obtained by cryo-electron microscopy (cryoEM) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / G-protein Coupled Receptor / 5-HT2AR / serotonin / psychedelics / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to cytoskeleton / positive regulation of heat generation / 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding / Gq/11-coupled serotonin receptor activity / positive regulation of phosphatidylinositol biosynthetic process / G protein-coupled serotonin receptor signaling pathway / G protein-coupled serotonin receptor complex / neurofilament / cell body fiber / phospholipase C-activating serotonin receptor signaling pathway ...protein localization to cytoskeleton / positive regulation of heat generation / 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding / Gq/11-coupled serotonin receptor activity / positive regulation of phosphatidylinositol biosynthetic process / G protein-coupled serotonin receptor signaling pathway / G protein-coupled serotonin receptor complex / neurofilament / cell body fiber / phospholipase C-activating serotonin receptor signaling pathway / artery smooth muscle contraction / positive regulation of cytokine production involved in immune response / Serotonin receptors / serotonin receptor activity / G protein-coupled serotonin receptor activity / serotonin receptor signaling pathway / sensitization / urinary bladder smooth muscle contraction / neurotransmitter receptor activity / serotonin binding / positive regulation of platelet aggregation / negative regulation of synaptic transmission, glutamatergic / positive regulation of DNA biosynthetic process / temperature homeostasis / detection of temperature stimulus involved in sensory perception of pain / regulation of dopamine secretion / negative regulation of potassium ion transport / protein tyrosine kinase activator activity / positive regulation of vasoconstriction / positive regulation of execution phase of apoptosis / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of fat cell differentiation / behavioral response to cocaine / release of sequestered calcium ion into cytosol / presynaptic modulation of chemical synaptic transmission / positive regulation of glycolytic process / dendritic shaft / glycolytic process / caveola / memory / intracellular calcium ion homeostasis / positive regulation of inflammatory response / positive regulation of neuron apoptotic process / positive regulation of cytosolic calcium ion concentration / virus receptor activity / presynaptic membrane / cytoplasmic vesicle / G alpha (q) signalling events / chemical synaptic transmission / postsynaptic membrane / positive regulation of ERK1 and ERK2 cascade / response to xenobiotic stimulus / axon / neuronal cell body / positive regulation of cell population proliferation / dendrite / protein-containing complex binding / glutamatergic synapse / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Gumpper RH / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: The structural diversity of psychedelic drug actions revealed. Authors: Ryan H Gumpper / Manish K Jain / Kuglae Kim / Renhong Sun / Ning Sun / Zhongli Xu / Jeffrey F DiBerto / Brian E Krumm / Nicholas J Kapolka / H Ümit Kaniskan / David E Nichols / Jian Jin / ...Authors: Ryan H Gumpper / Manish K Jain / Kuglae Kim / Renhong Sun / Ning Sun / Zhongli Xu / Jeffrey F DiBerto / Brian E Krumm / Nicholas J Kapolka / H Ümit Kaniskan / David E Nichols / Jian Jin / Jonathan F Fay / Bryan L Roth /   Abstract: There is currently a resurgence in exploring the utility of classical psychedelics to treat depression, addiction, anxiety disorders, cluster headaches, and many other neuropsychiatric disorders. A ...There is currently a resurgence in exploring the utility of classical psychedelics to treat depression, addiction, anxiety disorders, cluster headaches, and many other neuropsychiatric disorders. A biological target of these compounds, and a hypothesized target for their therapeutic actions, is the 5-HT serotonin receptor. Here, we present 7 cryo-EM structures covering all major compound classes of psychedelic and non-psychedelic agonists, including a β-arrestin-biased compound RS130-180. Identifying the molecular interactions between various psychedelics and the 5-HT receptor reveals both common and distinct motifs among the examined psychedelic chemotypes. These findings lead to a broader mechanistic understanding of 5-HT activation, which can catalyze the development of novel chemotypes with potential therapeutic utility and fewer side effects. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43797.map.gz emd_43797.map.gz | 109.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43797-v30.xml emd-43797-v30.xml emd-43797.xml emd-43797.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43797.png emd_43797.png | 35.7 KB | ||
| Masks |  emd_43797_msk_1.map emd_43797_msk_1.map | 115.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43797.cif.gz emd-43797.cif.gz | 6.2 KB | ||
| Others |  emd_43797_half_map_1.map.gz emd_43797_half_map_1.map.gz emd_43797_half_map_2.map.gz emd_43797_half_map_2.map.gz | 107.6 MB 107.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43797 http://ftp.pdbj.org/pub/emdb/structures/EMD-43797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43797 | HTTPS FTP |
-Related structure data
| Related structure data |  9arxMC  9aryC  9arzC  9as0C  9as1C  9as2C  9as3C  9as4C  9as5C  9as6C  9as7C  9as8C  9as9C  9asaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43797.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43797.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43797_msk_1.map emd_43797_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_43797_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43797_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 5-HT2AR in complex with mini-Gq
| Entire | Name: 5-HT2AR in complex with mini-Gq |
|---|---|
| Components |
|
-Supramolecule #1: 5-HT2AR in complex with mini-Gq
| Supramolecule | Name: 5-HT2AR in complex with mini-Gq / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 5-hydroxytryptamine receptor 2A
| Macromolecule | Name: 5-hydroxytryptamine receptor 2A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.648945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDILCEENTS LSSTTNSLMQ LNDDTRLYSN DFNSGEANTS DAFNWTVDSE NRTNLSCEGC LSPSCLSLLH LQEKNWSALL TAVVIILTI AGNILVIMAV SLEKKLQNAT NYFLMSLAIA DMLLGFLVMP VSMLTILYGY RWPLPSKLCA VWIYLDVLFS T ASIMHLCA ...String: MDILCEENTS LSSTTNSLMQ LNDDTRLYSN DFNSGEANTS DAFNWTVDSE NRTNLSCEGC LSPSCLSLLH LQEKNWSALL TAVVIILTI AGNILVIMAV SLEKKLQNAT NYFLMSLAIA DMLLGFLVMP VSMLTILYGY RWPLPSKLCA VWIYLDVLFS T ASIMHLCA ISLDRYVAIQ NPIHHSRFNS RTKAFLKIIA VWTISVGISM PIPVFGLQDD SKVFKEGSCL LADDNFVLIG SF VSFFIPL TIMVITYFLT IKSLQKEATL CVSDLGTRAK LASFSFLPQS SLSSEKLFQR SIHREPGSYT GRRTMQSISN EQK ACKVLG IVFFLFVVMW CPFFITNIMA VICKESCNED VIGALLNVFV WIGYLSSAVN PLVYTLFNKT YRSAFSRYIQ CQYK ENKKP LQLILVNTIP ALAYKSSQLQ MGQKKNSKQD AKTTDNDCSM VALGKQHSEE ASKDNSDGVN EKVSCV UniProtKB: 5-hydroxytryptamine receptor 2A |
-Macromolecule #2: SEROTONIN
| Macromolecule | Name: SEROTONIN / type: ligand / ID: 2 / Number of copies: 1 / Formula: SRO |
|---|---|
| Molecular weight | Theoretical: 176.215 Da |
| Chemical component information | 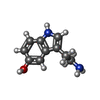 ChemComp-SRO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.16 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































