[English] 日本語
 Yorodumi
Yorodumi- EMDB-43198: Cryo-EM structure of FoxA1 and GATA4 in complex with H14 chromatosome -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of FoxA1 and GATA4 in complex with H14 chromatosome | |||||||||
 Map data Map data | Main class, linker DNA bended by FoxA1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / pioneer transcription factors / DNA binding proteins / transcription / chromatin / NUCLEAR PROTEIN / Linker histones / chromatosome / NUCLEAR PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationalveolar secondary septum development / respiratory basal cell differentiation / positive regulation of dopaminergic neuron differentiation / mesenchymal-epithelial cell signaling involved in prostate gland development / anatomical structure formation involved in morphogenesis / prostate gland stromal morphogenesis / positive regulation of cell-cell adhesion mediated by cadherin / epithelial cell maturation involved in prostate gland development / neuron fate specification / lung epithelial cell differentiation ...alveolar secondary septum development / respiratory basal cell differentiation / positive regulation of dopaminergic neuron differentiation / mesenchymal-epithelial cell signaling involved in prostate gland development / anatomical structure formation involved in morphogenesis / prostate gland stromal morphogenesis / positive regulation of cell-cell adhesion mediated by cadherin / epithelial cell maturation involved in prostate gland development / neuron fate specification / lung epithelial cell differentiation / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / dorsal/ventral neural tube patterning / prostate gland epithelium morphogenesis / negative regulation of DNA recombination / Formation of axial mesoderm / dopaminergic neuron differentiation / Apoptosis induced DNA fragmentation / positive regulation of smoothened signaling pathway / hormone metabolic process / chromosome condensation / negative regulation of epithelial to mesenchymal transition / nucleosomal DNA binding / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / smoothened signaling pathway / positive regulation of intracellular estrogen receptor signaling pathway / epithelial tube branching involved in lung morphogenesis / microvillus / anatomical structure morphogenesis / negative regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of DNA-binding transcription factor activity / negative regulation of megakaryocyte differentiation / heterochromatin / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / Notch signaling pathway / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / telomere organization / positive regulation of mitotic cell cycle / Interleukin-7 signaling / Inhibition of DNA recombination at telomere / RNA Polymerase I Promoter Opening / Meiotic synapsis / Assembly of the ORC complex at the origin of replication / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / epigenetic regulation of gene expression / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / innate immune response in mucosa / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / HDACs deacetylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / lipopolysaccharide binding / Transcriptional regulation by small RNAs / euchromatin / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / HDMs demethylate histones / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / chromatin DNA binding / DNA Damage/Telomere Stress Induced Senescence / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / positive regulation of miRNA transcription / Meiotic recombination / Pre-NOTCH Transcription and Translation / Metalloprotease DUBs / histone deacetylase binding / RMTs methylate histone arginines / Activation of anterior HOX genes in hindbrain development during early embryogenesis / fibrillar center / Transcriptional regulation of granulopoiesis / HCMV Early Events / sequence-specific double-stranded DNA binding / antimicrobial humoral immune response mediated by antimicrobial peptide / structural constituent of chromatin / UCH proteinases / antibacterial humoral response / response to estradiol / glucose homeostasis / nucleosome / heterochromatin formation / nucleosome assembly Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Zhou BR / Bai Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2024 Journal: Mol.Cell / Year: 2024Title: Cryo-EM structure of FoxA1 and GATA4 in complex with H14 chromatosome Authors: Zhou BR / Bai Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43198.map.gz emd_43198.map.gz | 58.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43198-v30.xml emd-43198-v30.xml emd-43198.xml emd-43198.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43198.png emd_43198.png | 130.8 KB | ||
| Filedesc metadata |  emd-43198.cif.gz emd-43198.cif.gz | 7.2 KB | ||
| Others |  emd_43198_additional_1.map.gz emd_43198_additional_1.map.gz emd_43198_half_map_1.map.gz emd_43198_half_map_1.map.gz emd_43198_half_map_2.map.gz emd_43198_half_map_2.map.gz | 58.6 MB 5.9 MB 5.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43198 http://ftp.pdbj.org/pub/emdb/structures/EMD-43198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43198 | HTTPS FTP |
-Validation report
| Summary document |  emd_43198_validation.pdf.gz emd_43198_validation.pdf.gz | 571.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43198_full_validation.pdf.gz emd_43198_full_validation.pdf.gz | 571.5 KB | Display | |
| Data in XML |  emd_43198_validation.xml.gz emd_43198_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_43198_validation.cif.gz emd_43198_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43198 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43198 | HTTPS FTP |
-Related structure data
| Related structure data |  8vg2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43198.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43198.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main class, linker DNA bended by FoxA1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||
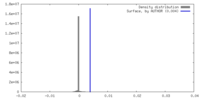
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Minor class, linker DNA less bent
| File | emd_43198_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Minor class, linker DNA less bent | ||||||||||||
| Projections & Slices |
| ||||||||||||
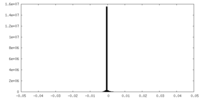
| Density Histograms |
-Half map: Half map 1
| File | emd_43198_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
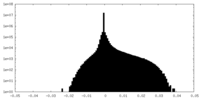
| Density Histograms |
-Half map: Half map 1
| File | emd_43198_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
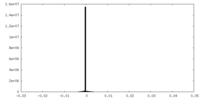
| Density Histograms |
- Sample components
Sample components
-Entire : FoxA1 and GATA4 in complex with H1.4 chromatosome
| Entire | Name: FoxA1 and GATA4 in complex with H1.4 chromatosome |
|---|---|
| Components |
|
-Supramolecule #1: FoxA1 and GATA4 in complex with H1.4 chromatosome
| Supramolecule | Name: FoxA1 and GATA4 in complex with H1.4 chromatosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.437167 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEACEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A type 1-B/E
| Macromolecule | Name: Histone H2A type 1-B/E / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.165551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A type 1-B/E |
-Macromolecule #4: Histone H2B type 1-J
| Macromolecule | Name: Histone H2B type 1-J / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.935239 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSI YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSAK UniProtKB: Histone H2B type 1-J |
-Macromolecule #7: Hepatocyte nuclear factor 3-alpha
| Macromolecule | Name: Hepatocyte nuclear factor 3-alpha / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.022562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGTVKMEGH ETSDWNSYYA DTQEAYSSVP VSNMNSGLGS MNSMNTYMTM NTMTTSGNMT PASFNMSYAN PGLGAGLSPG AVAGMPGGS AGAMNSMTAA GVTAMGTALS PSGMGAMGAQ QAASMNGLGP YAAAMNPCMS PMAYAPSNLG RSRAGGGGDA K TFKRSYPH ...String: MLGTVKMEGH ETSDWNSYYA DTQEAYSSVP VSNMNSGLGS MNSMNTYMTM NTMTTSGNMT PASFNMSYAN PGLGAGLSPG AVAGMPGGS AGAMNSMTAA GVTAMGTALS PSGMGAMGAQ QAASMNGLGP YAAAMNPCMS PMAYAPSNLG RSRAGGGGDA K TFKRSYPH AKPPYSYISL ITMAIQQAPS KMLTLSEIYQ WIMDLFPYYR QNQQRWQNSI RHSLSFNDCF VKVARSPDKP GK GSYWTLH PDSGNMFENG CYLRRQKRFK CEKQPGAGGG GGSGSGGSGA KGGPESRKDP SGASNPSADS PLHRGVHGKT GQL EGAPAP GPAASPQTLD HSGATATGGA SELKTPASST APPISSGPGA LASVPASHPA HGLAPHESQL HLKGDPHYSF NHPF SINNL MSSSEQQHKL DFKAYEQALQ YSPYGSTLPA SLPLGSASVT TRSPIEPSAL EPAYYQGVYS RPVLNTSHHH HHH UniProtKB: Hepatocyte nuclear factor 3-alpha |
-Macromolecule #8: Histone H1.4
| Macromolecule | Name: Histone H1.4 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.760404 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSETAPAAPA APAPAEKTPV KKKARKSAGA AKRKASGPPV SELITKAVAA SKERSGVSLA ALKKALAAAG YDVEKNNSRI KLGLKSLVS KGTLVQTKGT GASGSFKLNK KAASGEAKPK AKKAGAAKAK KPAGAAKKPK KATGAATPKK SAKKTPKKAK K PAAAAGAK ...String: MSETAPAAPA APAPAEKTPV KKKARKSAGA AKRKASGPPV SELITKAVAA SKERSGVSLA ALKKALAAAG YDVEKNNSRI KLGLKSLVS KGTLVQTKGT GASGSFKLNK KAASGEAKPK AKKAGAAKAK KPAGAAKKPK KATGAATPKK SAKKTPKKAK K PAAAAGAK KAKSPKKAKA AKPKKAPKSP AKAKAVKPKA AKPKTAKPKA AKPKKAAAKK KHHHHHH UniProtKB: Histone H1.4 |
-Macromolecule #5: DNA (196-MER)
| Macromolecule | Name: DNA (196-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 64.918344 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DG)(DA)(DG)(DA)(DT)(DG) (DG)(DT)(DA)(DC)(DT)(DT)(DT)(DG)(DT)(DG) (DT)(DC)(DT)(DC)(DC)(DT)(DG)(DC)(DT) (DC)(DT)(DG)(DT)(DC)(DA)(DG)(DC)(DA)(DG) (DG) (DG)(DC)(DA)(DC)(DT)(DG) ...String: (DA)(DT)(DC)(DC)(DG)(DA)(DG)(DA)(DT)(DG) (DG)(DT)(DA)(DC)(DT)(DT)(DT)(DG)(DT)(DG) (DT)(DC)(DT)(DC)(DC)(DT)(DG)(DC)(DT) (DC)(DT)(DG)(DT)(DC)(DA)(DG)(DC)(DA)(DG) (DG) (DG)(DC)(DA)(DC)(DT)(DG)(DT)(DA) (DC)(DT)(DT)(DG)(DC)(DT)(DG)(DA)(DT)(DA) (DC)(DC) (DA)(DG)(DG)(DG)(DA)(DA)(DT) (DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG) (DT)(DA)(DG) (DA)(DC)(DA)(DG)(DC)(DT) (DC)(DT)(DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC) (DT)(DT)(DA)(DA) (DA)(DC)(DG)(DC)(DA) (DC)(DG)(DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT) (DG)(DT)(DC)(DC)(DC) (DC)(DC)(DG)(DC) (DG)(DT)(DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG) (DC)(DC)(DA)(DA)(DG)(DG) (DG)(DG)(DA) (DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA) (DG)(DT)(DC)(DT)(DC)(DC)(DA) (DG)(DG) (DC)(DA)(DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG) (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DC) (DA) (DT)(DC)(DA)(DG)(DG)(DC)(DC)(DA)(DA)(DC) (DT)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DT)(DT)(DT)(DA)(DG)(DT)(DA)(DT)(DG)(DA) (DT) |
-Macromolecule #6: DNA (196-MER)
| Macromolecule | Name: DNA (196-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 65.368727 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DT)(DA)(DC)(DT)(DA)(DA) (DA)(DC)(DG)(DT)(DA)(DG)(DA)(DC)(DA)(DA) (DG)(DT)(DT)(DG)(DG)(DC)(DC)(DT)(DG) (DA)(DT)(DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DC) (DT)(DG)(DA)(DC)(DA)(DC) ...String: (DA)(DT)(DC)(DA)(DT)(DA)(DC)(DT)(DA)(DA) (DA)(DC)(DG)(DT)(DA)(DG)(DA)(DC)(DA)(DA) (DG)(DT)(DT)(DG)(DG)(DC)(DC)(DT)(DG) (DA)(DT)(DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DC) (DT)(DG)(DA)(DC)(DA)(DC)(DG)(DT) (DG)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC) (DT)(DA) (DG)(DG)(DG)(DA)(DG)(DT)(DA) (DA)(DT)(DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG) (DC)(DG)(DG) (DT)(DT)(DA)(DA)(DA)(DA) (DC)(DG)(DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC) (DA)(DG)(DC)(DG) (DC)(DG)(DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA) (DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA) (DC)(DG)(DA)(DC)(DC)(DA) (DA)(DT)(DT) (DG)(DA)(DT)(DT)(DC)(DC)(DC)(DT)(DG)(DG) (DT)(DA)(DT)(DC)(DA)(DG)(DC) (DA)(DA) (DG)(DT)(DA)(DC)(DA)(DG)(DT)(DG)(DC)(DC) (DC)(DT)(DG)(DC)(DT)(DG)(DA)(DC) (DA) (DG)(DA)(DG)(DC)(DA)(DG)(DG)(DA)(DG)(DA) (DC)(DA)(DC)(DA)(DA)(DA)(DG)(DT)(DA) (DC)(DC)(DA)(DT)(DC)(DT)(DC)(DG)(DG)(DA) (DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)