[English] 日本語
 Yorodumi
Yorodumi- EMDB-43034: Alpha7-nicotinic acetylcholine receptor time resolved bound to ep... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ION CHANNEL / MEMBRANE PROTEIN / NICOTINIC RECEPTOR | |||||||||
| Function / homology |  Function and homology information Function and homology informationsensory processing / synaptic transmission involved in micturition / dendrite arborization / response to acetylcholine / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / acetylcholine receptor activity / acetylcholine-gated channel complex / regulation of amyloid fibril formation / chloride channel regulator activity / acetylcholine-gated monoatomic cation-selective channel activity ...sensory processing / synaptic transmission involved in micturition / dendrite arborization / response to acetylcholine / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / acetylcholine receptor activity / acetylcholine-gated channel complex / regulation of amyloid fibril formation / chloride channel regulator activity / acetylcholine-gated monoatomic cation-selective channel activity / short-term memory / cation channel complex / acetylcholine binding / dendritic spine organization / regulation of amyloid precursor protein catabolic process / acetylcholine receptor signaling pathway / neurotransmitter receptor complex / positive regulation of amyloid-beta formation / positive regulation of protein metabolic process / negative regulation of amyloid-beta formation / response to amyloid-beta / ligand-gated ion channel signaling pathway / negative regulation of tumor necrosis factor production / modulation of excitatory postsynaptic potential / plasma membrane raft / monoatomic ion channel activity / toxic substance binding / monoatomic ion transport / negative regulation of cytokine production involved in inflammatory response / negative regulation of canonical NF-kappaB signal transduction / positive regulation of excitatory postsynaptic potential / positive regulation of long-term synaptic potentiation / response to nicotine / excitatory postsynaptic potential / regulation of membrane potential / electron transport chain / synapse organization / cognition / calcium channel activity / memory / positive regulation of angiogenesis / intracellular calcium ion homeostasis / calcium ion transport / transmembrane signaling receptor activity / amyloid-beta binding / monoatomic ion transmembrane transport / chemical synaptic transmission / response to hypoxia / learning or memory / postsynaptic membrane / electron transfer activity / periplasmic space / positive regulation of ERK1 and ERK2 cascade / positive regulation of MAPK cascade / neuron projection / postsynapse / iron ion binding / heme binding / positive regulation of cell population proliferation / dendrite / synapse / endoplasmic reticulum membrane / signal transduction / protein homodimerization activity / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
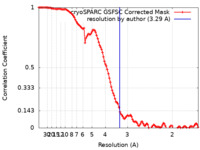
| Method | single particle reconstruction / cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Burke SM / Noviello CM / Hibbs RE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Structural mechanisms of α7 nicotinic receptor allosteric modulation and activation. Authors: Sean M Burke / Mariia Avstrikova / Colleen M Noviello / Nuriya Mukhtasimova / Jean-Pierre Changeux / Ganesh A Thakur / Steven M Sine / Marco Cecchini / Ryan E Hibbs /   Abstract: The α7 nicotinic acetylcholine receptor is a pentameric ligand-gated ion channel that plays an important role in cholinergic signaling throughout the nervous system. Its unique physiological ...The α7 nicotinic acetylcholine receptor is a pentameric ligand-gated ion channel that plays an important role in cholinergic signaling throughout the nervous system. Its unique physiological characteristics and implications in neurological disorders and inflammation make it a promising but challenging therapeutic target. Positive allosteric modulators overcome limitations of traditional α7 agonists, but their potentiation mechanisms remain unclear. Here, we present high-resolution structures of α7-modulator complexes, revealing partially overlapping binding sites but varying conformational states. Structure-guided functional and computational tests suggest that differences in modulator activity arise from the stable rotation of a channel gating residue out of the pore. We extend the study using a time-resolved cryoelectron microscopy (cryo-EM) approach to reveal asymmetric state transitions for this homomeric channel and also find that a modulator with allosteric agonist activity exploits a distinct channel-gating mechanism. These results define mechanisms of α7 allosteric modulation and activation with implications across the pentameric receptor superfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43034.map.gz emd_43034.map.gz | 136.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43034-v30.xml emd-43034-v30.xml emd-43034.xml emd-43034.xml | 42.4 KB 42.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43034_fsc.xml emd_43034_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_43034.png emd_43034.png | 57.2 KB | ||
| Filedesc metadata |  emd-43034.cif.gz emd-43034.cif.gz | 8.2 KB | ||
| Others |  emd_43034_additional_1.map.gz emd_43034_additional_1.map.gz emd_43034_additional_2.map.gz emd_43034_additional_2.map.gz emd_43034_additional_3.map.gz emd_43034_additional_3.map.gz emd_43034_additional_4.map.gz emd_43034_additional_4.map.gz emd_43034_additional_5.map.gz emd_43034_additional_5.map.gz emd_43034_additional_6.map.gz emd_43034_additional_6.map.gz emd_43034_additional_7.map.gz emd_43034_additional_7.map.gz emd_43034_additional_8.map.gz emd_43034_additional_8.map.gz emd_43034_additional_9.map.gz emd_43034_additional_9.map.gz emd_43034_half_map_1.map.gz emd_43034_half_map_1.map.gz emd_43034_half_map_2.map.gz emd_43034_half_map_2.map.gz | 72.2 MB 136.8 MB 136.8 MB 72.3 MB 134.3 MB 134.1 MB 134.2 MB 69.2 MB 134.2 MB 134.1 MB 134.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43034 http://ftp.pdbj.org/pub/emdb/structures/EMD-43034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43034 | HTTPS FTP |
-Related structure data
| Related structure data |  8v8cMC  8ut1C  8utbC  8uzjC  8v80C  8v82C  8v86C  8v88C  8v89C  8v8aC  8v8dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43034.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43034.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
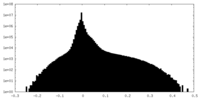
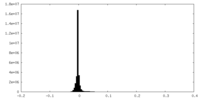
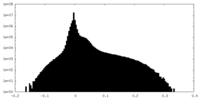
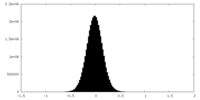
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: #9
+Additional map: #8
+Additional map: #7
+Additional map: #6
+Additional map: #5
+Additional map: #4
+Additional map: #3
+Additional map: #2
+Additional map: #1
+Half map: #2
+Half map: #1
- Sample components
Sample components
-Entire : Alpha7-nicotinic acetylcholine receptor time resolved bound to ep...
| Entire | Name: Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 |
|---|---|
| Components |
|
-Supramolecule #1: Alpha7-nicotinic acetylcholine receptor time resolved bound to ep...
| Supramolecule | Name: Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Neuronal acetylcholine receptor subunit alpha-7,Soluble cytochrom...
| Macromolecule | Name: Neuronal acetylcholine receptor subunit alpha-7,Soluble cytochrome b562 type: protein_or_peptide / ID: 1 Details: 351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage) ...Details: 351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage),351-353 (linker) 354-460 (SOLUBLE CYTOCHROME B562 FUSION) 551-558 (strep tag II) 559-571 (linker) 572-579 (strep tag II) 580-582 (linker) 583-599 (T2A self cleaving peptide post cleavage) Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.220797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EFQRKLYKEL VKNYNPLERP VANDSQPLTV YFSLSLLQIM DVDEKNQVLT TNIWLQMSWT DHYLQWNVSE YPGVKTVRFP DGQIWKPDI LLYNSADERF DATFHTNVLV NSSGHCQYLP PGIFKSSCYI DVRWFPFDVQ HCKLKFGSWS YGGWSLDLQM Q EADISGYI ...String: EFQRKLYKEL VKNYNPLERP VANDSQPLTV YFSLSLLQIM DVDEKNQVLT TNIWLQMSWT DHYLQWNVSE YPGVKTVRFP DGQIWKPDI LLYNSADERF DATFHTNVLV NSSGHCQYLP PGIFKSSCYI DVRWFPFDVQ HCKLKFGSWS YGGWSLDLQM Q EADISGYI PNGEWDLVGI PGKRSERFYE CCKEPYPDVT FTVTMRRRTL YYGLNLLIPC VLISALALLV FLLPADSGEK IS LGITVLL SLTVFMLLVA EIMPATSDSV PLIAQYFAST MIIVGLSVVV TVIVLQYHHH DPDGGKMPKW TRVILLNWCA WFL RMKRPG EDKVRPACQH KQRRCSLACG RMACAGAMAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPP KLEDK SPDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLSPTHD EHLLHGGQPP EGDPD LAKI LEEVRYIANR FRCQDESEAV CSEWKFAACV VDRLCLMAFS VFTIICTIGI LMSAPNFVEA VSKDFAWSHP QFEKGG GSG GGSGGSSAWS HPQFEKGSGE GRGSLLTCGD VEENPG UniProtKB: Neuronal acetylcholine receptor subunit alpha-7, Soluble cytochrome b562 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: EPIBATIDINE
| Macromolecule | Name: EPIBATIDINE / type: ligand / ID: 4 / Number of copies: 5 / Formula: EPJ |
|---|---|
| Molecular weight | Theoretical: 208.687 Da |
| Chemical component information |  ChemComp-EPJ: |
-Macromolecule #5: N-(5-Chloro-2,4-dimethoxyphenyl)-N'-(5-methyl-3-isoxazolyl)-urea
| Macromolecule | Name: N-(5-Chloro-2,4-dimethoxyphenyl)-N'-(5-methyl-3-isoxazolyl)-urea type: ligand / ID: 5 / Number of copies: 5 / Formula: I34 |
|---|---|
| Molecular weight | Theoretical: 311.721 Da |
| Chemical component information | 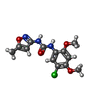 ChemComp-I34: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)