[English] 日本語
 Yorodumi
Yorodumi- EMDB-42983: Structure of the Human Respirovirus 3 Fusion Protein Bound to Cam... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Human Respirovirus 3 Fusion Protein Bound to Camelid Nanobodies 4C03 and 4C06 | |||||||||
 Map data Map data | Final refinement volume. DeepEMhancer sharpened. Used for building coordinates. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral fusion protein / camelid nanobodies / viral glycoprotein / membrane fusion / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | |||||||||
| Biological species |  Human respirovirus 3 / Human respirovirus 3 /  | |||||||||
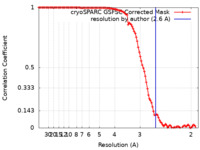
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Johnson NV / Ramamohan AR / McLellan JS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for potent neutralization of human respirovirus type 3 by protective single-domain camelid antibodies. Authors: Nicole V Johnson / Revina C van Scherpenzeel / Mark J G Bakkers / Ajit R Ramamohan / Daan van Overveld / Lam Le / Johannes P M Langedijk / Joost A Kolkman / Jason S McLellan /    Abstract: Respirovirus 3 is a leading cause of severe acute respiratory infections in vulnerable human populations. Entry into host cells is facilitated by the attachment glycoprotein and the fusion ...Respirovirus 3 is a leading cause of severe acute respiratory infections in vulnerable human populations. Entry into host cells is facilitated by the attachment glycoprotein and the fusion glycoprotein (F). Because of its crucial role, F represents an attractive therapeutic target. Here, we identify 13 F-directed heavy-chain-only antibody fragments that neutralize recombinant respirovirus 3. High-resolution cryo-EM structures of antibody fragments bound to the prefusion conformation of F reveal three distinct, previously uncharacterized epitopes. All three antibody fragments bind quaternary epitopes on F, suggesting mechanisms for neutralization that may include stabilization of the prefusion conformation. Studies in cotton rats demonstrate the prophylactic efficacy of these antibody fragments in reducing viral load in the lungs and nasal passages. These data highlight the potential of heavy-chain-only antibody fragments as effective interventions against respirovirus 3 infection and identify neutralizing epitopes that can be targeted for therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42983.map.gz emd_42983.map.gz | 105.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42983-v30.xml emd-42983-v30.xml emd-42983.xml emd-42983.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42983_fsc.xml emd_42983_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42983.png emd_42983.png | 101.1 KB | ||
| Filedesc metadata |  emd-42983.cif.gz emd-42983.cif.gz | 6.1 KB | ||
| Others |  emd_42983_additional_1.map.gz emd_42983_additional_1.map.gz emd_42983_half_map_1.map.gz emd_42983_half_map_1.map.gz emd_42983_half_map_2.map.gz emd_42983_half_map_2.map.gz | 117.9 MB 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42983 http://ftp.pdbj.org/pub/emdb/structures/EMD-42983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42983 | HTTPS FTP |
-Related structure data
| Related structure data |  8v5kMC  8v62C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refinement volume. DeepEMhancer sharpened. Used for building coordinates. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Final refined volume. Sharpened map. Not DeepEMhancer sharpened.
| File | emd_42983_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refined volume. Sharpened map. Not DeepEMhancer sharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
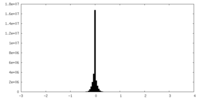
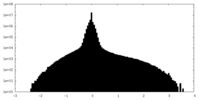
| Density Histograms |
-Half map: half map B
| File | emd_42983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
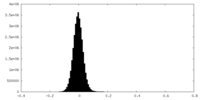
| Density Histograms |
-Half map: half map A
| File | emd_42983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
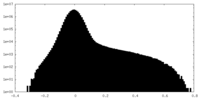
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion Human Respirovirus 3 Fusion Protein bound to Camelid na...
| Entire | Name: Prefusion Human Respirovirus 3 Fusion Protein bound to Camelid nanobodies 4C03 and 4C06 |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion Human Respirovirus 3 Fusion Protein bound to Camelid na...
| Supramolecule | Name: Prefusion Human Respirovirus 3 Fusion Protein bound to Camelid nanobodies 4C03 and 4C06 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
| Molecular weight | Theoretical: 277 KDa |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
| Molecular weight | Theoretical: 55.364965 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QIDITKLQHV GVLVNSPKGM KISQNFETRY LILSLIPKIE DSNSCGDQQI KQYKRLLDRL IIPLYDGLRL QKDVIVTNQE SNENTDPRT ERFFGGVIGT IALGVATSAQ ITAAVALVEA KQARSDIEKL KEAIRDTNKA VQSVQSSPGN LIVAIKSVQD Y VNKEIVPC ...String: QIDITKLQHV GVLVNSPKGM KISQNFETRY LILSLIPKIE DSNSCGDQQI KQYKRLLDRL IIPLYDGLRL QKDVIVTNQE SNENTDPRT ERFFGGVIGT IALGVATSAQ ITAAVALVEA KQARSDIEKL KEAIRDTNKA VQSVQSSPGN LIVAIKSVQD Y VNKEIVPC IARLGCEACG LLLGLALDQH YSELTNIFGD NIGSLQEKGI KLQGIASLYR TNITEIFTTS TVDKYDIYDL LF TESIKVR VIDVDLNDYS ITLQVRLPLL TRLLNTQIYK VDSISYNIQN REWYIPLPSH IMTKGAFLGG ADVKECIEAF SSY ICPSDP GFVLNHEMES CLSGNISQCP RTTVTSDIVP RYAFVNGGVV ANCITTTCTC NGIGNRINQP PDQGVKIITH KECN TIGIN GMLFNTNKEG TLAFYTPDDI TLNNSVALNP IDISIELNKA KSDLEESKEW IRRSNQKLDS IEDKIEEILS KIYHI ENEI ARIKKLIGEA EPEA UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: Camelid Nanobody 4C03
| Macromolecule | Name: Camelid Nanobody 4C03 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.726373 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVRAGGSLRL SCAASLRDLH TRTFYMGWFR QDPGKEREFV AAIDWNTGAA SYPDSVKGRF TISKDNARNA VYLQMNNLK PEDTAVYYCA VGRPPLNRPT LAYYWGQGTQ VTVSS |
-Macromolecule #3: Camelid Nanobody 4C06
| Macromolecule | Name: Camelid Nanobody 4C06 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.487816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCSASGSLST IKALGWYRRA PGRERELVAS ITSAGETNYA DSAKGRFTVS TDNAKNTVDL RMNSLKPED TAVYYCYAES FVLNIYWGQG TQVTVSSG |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)