+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of AriA-AriB complex (Form III) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial Defense system / Toxin-antitoxin system / AriAB / PARIS / IMMUNE SYSTEM | |||||||||
| Function / homology | : / :  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
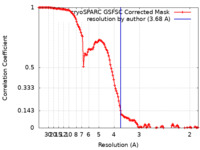
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Deep A / Corbett KD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Architecture and activation mechanism of the bacterial PARIS defence system. Authors: Amar Deep / Qishan Liang / Eray Enustun / Joe Pogliano / Kevin D Corbett /  Abstract: Bacteria and their viruses (bacteriophages or phages) are engaged in an intense evolutionary arms race. While the mechanisms of many bacterial antiphage defence systems are known, how these systems ...Bacteria and their viruses (bacteriophages or phages) are engaged in an intense evolutionary arms race. While the mechanisms of many bacterial antiphage defence systems are known, how these systems avoid toxicity outside infection yet activate quickly after infection is less well understood. Here we show that the bacterial phage anti-restriction-induced system (PARIS) operates as a toxin-antitoxin system, in which the antitoxin AriA sequesters and inactivates the toxin AriB until triggered by the T7 phage counterdefence protein Ocr. Using cryo-electron microscopy, we show that AriA is related to SMC-family ATPases but assembles into a distinctive homohexameric complex through two oligomerization interfaces. In uninfected cells, the AriA hexamer binds to up to three monomers of AriB, maintaining them in an inactive state. After Ocr binding, the AriA hexamer undergoes a structural rearrangement, releasing AriB and allowing it to dimerize and activate. AriB is a toprim/OLD-family nuclease, the activation of which arrests cell growth and inhibits phage propagation by globally inhibiting protein translation through specific cleavage of a lysine tRNA. Collectively, our findings reveal the intricate molecular mechanisms of a bacterial defence system triggered by a phage counterdefence protein, and highlight how an SMC-family ATPase has been adapted as a bacterial infection sensor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42968.map.gz emd_42968.map.gz | 107.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42968-v30.xml emd-42968-v30.xml emd-42968.xml emd-42968.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42968_fsc.xml emd_42968_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42968.png emd_42968.png | 130.1 KB | ||
| Filedesc metadata |  emd-42968.cif.gz emd-42968.cif.gz | 6.1 KB | ||
| Others |  emd_42968_half_map_1.map.gz emd_42968_half_map_1.map.gz emd_42968_half_map_2.map.gz emd_42968_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42968 http://ftp.pdbj.org/pub/emdb/structures/EMD-42968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42968 | HTTPS FTP |
-Related structure data
| Related structure data |  8v48MC  8v45C  8v46C  8v47C  8v49C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42968.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42968.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42968_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
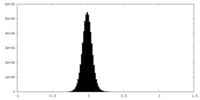
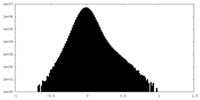
| Density Histograms |
-Half map: #1
| File | emd_42968_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
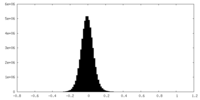
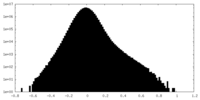
| Density Histograms |
- Sample components
Sample components
-Entire : AriAB complex
| Entire | Name: AriAB complex |
|---|---|
| Components |
|
-Supramolecule #1: AriAB complex
| Supramolecule | Name: AriAB complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: AriA-AriB |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 390 KDa |
-Macromolecule #1: AriA antitoxin
| Macromolecule | Name: AriA antitoxin / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.287113 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VAIRTISKIE LSKIHNRYNL TVDFFNDLNV IHGKNGAGKS TLIHVIANIV NGDFIRFAFL IFEEIKATYS DGLKIVIRRD KIDEQSFIS VTLSNGKYIK FAVGEAMATV REIESERHLR ERDVKSMLAM DIDKFVKENE LQKVRASYFP AFRTMLEAWS S SSDVGYER ...String: VAIRTISKIE LSKIHNRYNL TVDFFNDLNV IHGKNGAGKS TLIHVIANIV NGDFIRFAFL IFEEIKATYS DGLKIVIRRD KIDEQSFIS VTLSNGKYIK FAVGEAMATV REIESERHLR ERDVKSMLAM DIDKFVKENE LQKVRASYFP AFRTMLEAWS S SSDVGYER RVIRSSFYNR KASAFARELF GQFLPSINYP SPMEIEDRLR EEIRRAQLGI AAYESRTFSE SFVKVFSALF DN SSVEGEI TGELLKEIEG LAIAQDSSIK NGYYAEYSKV YEEIRSLINR NLKGKVENSV SGALVVYRDA LRDRQDYQEK AFS EIDNYM SSVNSFLEDK EMAYDFDLRR KYPKVGLKFP DGSWSPIRVL SSGERQLLTM LYAASKMGDD AIVLIDQPEI SLHI DWQED LLKRMLSQLS GRQIIVCTHS PSIATGYEDF MINISPEFIS SRDNDNHKDS EEMEEDESL UniProtKB: UNIPROTKB: D6IC77 |
-Macromolecule #2: AriB
| Macromolecule | Name: AriB / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.749578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSCAYTIDS YITLLTMSSK KRLLVEGRHD RSHLYQLIYK FNPASKVKID TAQDIKASDK AMSKNNRLKI ETIHSKVKGK DNISFLCDR AFREFAFNDQ IEDLLNSHYC DDSLYWTLGH SLENYFFNPS IIIDAFQFLS PSEYKYKAIE LFSELISSSF A VLAAVSLA ...String: MSSCAYTIDS YITLLTMSSK KRLLVEGRHD RSHLYQLIYK FNPASKVKID TAQDIKASDK AMSKNNRLKI ETIHSKVKGK DNISFLCDR AFREFAFNDQ IEDLLNSHYC DDSLYWTLGH SLENYFFNPS IIIDAFQFLS PSEYKYKAIE LFSELISSSF A VLAAVSLA AKDIDKAGLP AALIDWKDIV INDGTIKLIR RDSYDIDSAC VDSFFNAFDA VLPRVIASDV GICSRVVRGH TG ILLLQKL FSACLYYVGR EDDALQADSS ANYFCNLSEL SLTTALAESW VRKIGVLEDV YFPDSLLKNI E UniProtKB: UNIPROTKB: D6IC76 |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 5058 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8v48: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)