[English] 日本語
 Yorodumi
Yorodumi- EMDB-42943: Structure of the human systemic RNAi defective transmembrane prot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human systemic RNAi defective transmembrane protein 1 (hSIDT1) | |||||||||
 Map data Map data | Sharpened map resulting from local refinement in cryosparc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA interference / transmembrane protein / cholesterol or dsRNA uptake family. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA transmembrane transporter activity / RNA transport / NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / cholesterol binding / bioluminescence / generation of precursor metabolites and energy / virion component / double-stranded RNA binding / host cell cytoplasm ...RNA transmembrane transporter activity / RNA transport / NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / cholesterol binding / bioluminescence / generation of precursor metabolites and energy / virion component / double-stranded RNA binding / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / lysosome / hydrolase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
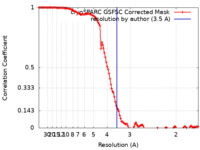
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Navratna V / Kumar A / Rana JK / Mosalaganti S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Structure of the human systemic RNAi defective transmembrane protein 1 (hSIDT1) reveals the conformational flexibility of its lipid binding domain. Authors: Vikas Navratna / Arvind Kumar / Jaimin K Rana / Shyamal Mosalaganti /  Abstract: In , inter-cellular transport of the small non-coding RNA causing systemic RNA interference (RNAi) is mediated by the transmembrane protein SID1, encoded by the gene in the systemic RNA interference- ...In , inter-cellular transport of the small non-coding RNA causing systemic RNA interference (RNAi) is mediated by the transmembrane protein SID1, encoded by the gene in the systemic RNA interference-defective () loci. SID1 shares structural and sequence similarity with cholesterol uptake protein 1 (CHUP1) and is classified as a member of the cholesterol uptake family (ChUP). Although systemic RNAi is not an evolutionarily conserved process, the gene products are found across the animal kingdom, suggesting the existence of other novel gene regulatory mechanisms mediated by small non-coding RNAs. Human homologs of gene products - hSIDT1 and hSIDT2 - mediate contact-dependent lipophilic small non-coding dsRNA transport. Here, we report the structure of recombinant human SIDT1. We find that the extra-cytosolic domain (ECD) of hSIDT1 adopts a double jelly roll fold, and the transmembrane domain (TMD) exists as two modules - a flexible lipid binding domain (LBD) and a rigid TMD core. Our structural analyses provide insights into the inherent conformational dynamics within the lipid binding domain in cholesterol uptake (ChUP) family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42943.map.gz emd_42943.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42943-v30.xml emd-42943-v30.xml emd-42943.xml emd-42943.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42943_fsc.xml emd_42943_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_42943.png emd_42943.png | 91.2 KB | ||
| Filedesc metadata |  emd-42943.cif.gz emd-42943.cif.gz | 7.2 KB | ||
| Others |  emd_42943_additional_1.map.gz emd_42943_additional_1.map.gz emd_42943_additional_2.map.gz emd_42943_additional_2.map.gz emd_42943_half_map_1.map.gz emd_42943_half_map_1.map.gz emd_42943_half_map_2.map.gz emd_42943_half_map_2.map.gz | 88.2 MB 994 KB 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42943 http://ftp.pdbj.org/pub/emdb/structures/EMD-42943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42943 | HTTPS FTP |
-Validation report
| Summary document |  emd_42943_validation.pdf.gz emd_42943_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42943_full_validation.pdf.gz emd_42943_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42943_validation.xml.gz emd_42943_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_42943_validation.cif.gz emd_42943_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42943 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42943 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42943 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42943 | HTTPS FTP |
-Related structure data
| Related structure data |  8v38MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42943.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42943.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map resulting from local refinement in cryosparc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
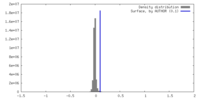
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: UnSharpened map resulting from local refinement in cryosparc
| File | emd_42943_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | UnSharpened map resulting from local refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
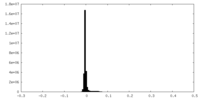
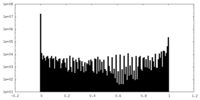
| Density Histograms |
-Additional map: Mask Used for refinement during local refinement refinement...
| File | emd_42943_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mask Used for refinement during local refinement refinement of symmetry expanded particles in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
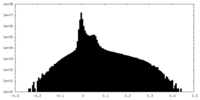
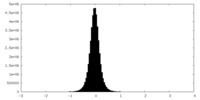
| Density Histograms |
-Half map: Half map resulting from Local refinement in cryosparc
| File | emd_42943_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map resulting from Local refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
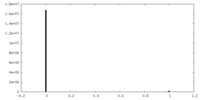
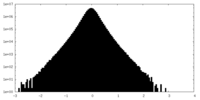
| Density Histograms |
-Half map: Half map resulting from Local refinement in cryosparc
| File | emd_42943_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map resulting from Local refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
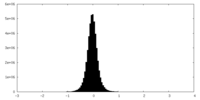
| Density Histograms |
- Sample components
Sample components
-Entire : Human systemic RNAi defective transmembrane protein 1 (hSIDT1)
| Entire | Name: Human systemic RNAi defective transmembrane protein 1 (hSIDT1) |
|---|---|
| Components |
|
-Supramolecule #1: Human systemic RNAi defective transmembrane protein 1 (hSIDT1)
| Supramolecule | Name: Human systemic RNAi defective transmembrane protein 1 (hSIDT1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Full length human SIDT1 expressed as a recombinant fusion protein with N-terminal GFP, in mammalian cells |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Lysosomes / Location in cell: Lysosomal membrane Homo sapiens (human) / Organelle: Lysosomes / Location in cell: Lysosomal membrane |
| Molecular weight | Theoretical: 123.9 KDa |
-Macromolecule #1: SID1 transmembrane family member 1,RNA-directed RNA polymerase L
| Macromolecule | Name: SID1 transmembrane family member 1,RNA-directed RNA polymerase L type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: NNS virus cap methyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.098141 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRGCLRLALL CALPWLLLAA SPGHPAKSPR QPPAPRRDPF DAARGADFDH VYSGVVNLST ENIYSFNYTS QPDQVTAVRV YVNSSSENL NYPVLVVVRQ QKEVLSWQVP LLFQGLYQRS YNYQEVSRTL CPSEATNETG PLQQLIFVDV ASMAPLGAQY K LLVTKLKH ...String: MRGCLRLALL CALPWLLLAA SPGHPAKSPR QPPAPRRDPF DAARGADFDH VYSGVVNLST ENIYSFNYTS QPDQVTAVRV YVNSSSENL NYPVLVVVRQ QKEVLSWQVP LLFQGLYQRS YNYQEVSRTL CPSEATNETG PLQQLIFVDV ASMAPLGAQY K LLVTKLKH FQLRTNVAFH FTASPSQPQY FLYKFPKDVD SVIIKVVSEM AYPCSVVSVQ NIMCPVYDLD HNVEFNGVYQ SM TKKAAIT LQKKDFPGEQ FFVVFVIKPE DYACGGSFFI QEKENQTWNL QRKKNLEVTI VPSIKESVYV KSSLFSVFIF LSF YLGCLL VGFVHYLRFQ RKSIDGSFGS NDGSGNMVAS HPIAASTPEG SNYGTIDESS SSPGRQMSSS DGGPPGQSDT DSSV EESDF DTMPDIESDK NIIRTKMFLY LSDLSRKDRR IVSKKYKIYF WNIITIAVFY ALPVIQLVIT YQTVVNVTGN QDICY YNFL CAHPLGVLSA FNNILSNLGH VLLGFLFLLI VLRRDILHRR ALEAKDIFAV EYGIPKHFGL FYAMGIALMM EGVLSA CYH VCPNYSNFQF DTSFMYMIAG LCMLKLYQTR HPDINASAYS AYASFAVVIM VTVLGVVFGK NDVWFWVIFS AIHVLAS LA LSTQIYYMGR FKIDLGIFRR AAMVFYTDCI QQCSRPLYMD RMVLLVVGNL VNWSFALFGL IYRPRDFASY MLGIFICN L LLYLAFYIIM KLRSSEKVLP VPLFCIVATA VMWAAALYFF FQNLSSWEGT PAESREKNRE CILLDFFDDH DIWHFLSAT ALFFSFLVLL TLDDDLDVVR RDQIPVFAAA FESRLEVLFQ GPAAAAVSKG EELFTGVVPI LVELDGDVNG HKFSVSGEGE GDATYGKLT LKFICTTGKL PVPWPTLVTT LTYGVQCFSR YPDHMKQHDF FKSAMPEGYV QERTIFFKDD GNYKTRAEVK F EGDTLVNR IELKGIDFKE DGNILGHKLE YNYNSHNVYI MADKQKNGIK VNFKIRHNIE DGSVQLADHY QQNTPIGDGP VL LPDNHYL STQSKLSKDP NEKRDHMVLL EFVTAAGITL GMDELYKSGG SGWSHPQFEK UniProtKB: SID1 transmembrane family member 1, RNA-directed RNA polymerase L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.026000000000000002 kPa / Details: 15 mA current | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | hSIDT1 in digitonin micelle, purified by Strep-Tactin affinity chromatography |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 17000 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: Made using Model angelo |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-8v38: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)