[English] 日本語
 Yorodumi
Yorodumi- EMDB-42891: Herpes simplex virus 1 polymerase W781V mutant holoenzyme bound t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herpes simplex virus 1 polymerase W781V mutant holoenzyme bound to DNA in editing conformation | |||||||||
 Map data Map data | sharpened map after polishing | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | herpes simplex virus / replication / DNA polymerase holoenzyme / W781V / editing conformation / TRANSFERASE-DNA complex / VIRAL PROTEIN | |||||||||
| Function / homology | : / :  Function and homology information Function and homology information | |||||||||
| Biological species |   Human herpesvirus 1 (strain KOS) / Human herpesvirus 1 (strain KOS) /   Human alphaherpesvirus 1 strain KOS / synthetic construct (others) Human alphaherpesvirus 1 strain KOS / synthetic construct (others) | |||||||||
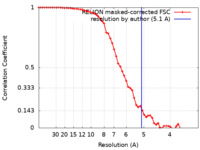
| Method | single particle reconstruction / cryo EM / Resolution: 5.1 Å | |||||||||
 Authors Authors | Pan J / Abraham J / Coen DM / Shankar S / Yang P / Hogle J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance. Authors: Sundaresh Shankar / Junhua Pan / Pan Yang / Yuemin Bian / Gábor Oroszlán / Zishuo Yu / Purba Mukherjee / David J Filman / James M Hogle / Mrinal Shekhar / Donald M Coen / Jonathan Abraham /    Abstract: DNA polymerases are important drug targets, and many structural studies have captured them in distinct conformations. However, a detailed understanding of the impact of polymerase conformational ...DNA polymerases are important drug targets, and many structural studies have captured them in distinct conformations. However, a detailed understanding of the impact of polymerase conformational dynamics on drug resistance is lacking. We determined cryoelectron microscopy (cryo-EM) structures of DNA-bound herpes simplex virus polymerase holoenzyme in multiple conformations and interacting with antivirals in clinical use. These structures reveal how the catalytic subunit Pol and the processivity factor UL42 bind DNA to promote processive DNA synthesis. Unexpectedly, in the absence of an incoming nucleotide, we observed Pol in multiple conformations with the closed state sampled by the fingers domain. Drug-bound structures reveal how antivirals may selectively bind enzymes that more readily adopt the closed conformation. Molecular dynamics simulations and the cryo-EM structure of a drug-resistant mutant indicate that some resistance mutations modulate conformational dynamics rather than directly impacting drug binding, thus clarifying mechanisms that drive drug selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42891.map.gz emd_42891.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42891-v30.xml emd-42891-v30.xml emd-42891.xml emd-42891.xml | 30.8 KB 30.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42891_fsc.xml emd_42891_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42891.png emd_42891.png | 38.8 KB | ||
| Filedesc metadata |  emd-42891.cif.gz emd-42891.cif.gz | 7.3 KB | ||
| Others |  emd_42891_additional_1.map.gz emd_42891_additional_1.map.gz emd_42891_additional_2.map.gz emd_42891_additional_2.map.gz emd_42891_additional_3.map.gz emd_42891_additional_3.map.gz emd_42891_half_map_1.map.gz emd_42891_half_map_1.map.gz emd_42891_half_map_2.map.gz emd_42891_half_map_2.map.gz | 13.9 MB 14 MB 11.8 MB 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42891 http://ftp.pdbj.org/pub/emdb/structures/EMD-42891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42891 | HTTPS FTP |
-Related structure data
| Related structure data |  8exxC  8v1qC  8v1rC  8v1sC  8v1tC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42891.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42891.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map after polishing | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.65 Å | ||||||||||||||||||||||||||||||||||||
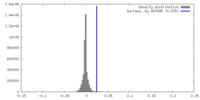
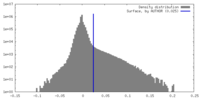
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: deepemhanced map prior to polishing
| File | emd_42891_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepemhanced map prior to polishing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: deepemhanced map after polishing
| File | emd_42891_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepemhanced map after polishing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unfiltered map after polishing
| File | emd_42891_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map after polishing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map 2 after polishing
| File | emd_42891_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 2 after polishing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map 1 after polishing
| File | emd_42891_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 1 after polishing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HSV-1 DNA polymerase W781V mutant holoenzyme in editing complex
| Entire | Name: HSV-1 DNA polymerase W781V mutant holoenzyme in editing complex |
|---|---|
| Components |
|
-Supramolecule #1: HSV-1 DNA polymerase W781V mutant holoenzyme in editing complex
| Supramolecule | Name: HSV-1 DNA polymerase W781V mutant holoenzyme in editing complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Herpes simplex virus type 1 KOS strain polymerase W781V mutant holoenzyme UL30W781V:UL42 in complex with primer DNA that has a 3'-end mismatch with template DNA |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 (strain KOS) Human herpesvirus 1 (strain KOS) |
| Molecular weight | Theoretical: 197 KDa |
-Macromolecule #1: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 1 Details: Herpes simplex virus type 1 (KOS strain) DNA polymerase UL30 with its N-terminal 42 residues deleted and replaced by an N-terminal poly-histidine tag in the expression construct Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS / Strain: KOS Human alphaherpesvirus 1 strain KOS / Strain: KOS |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHNFYN PYLAPVGTQQ KPTGPTQRHT YYSECDEFRF IAPRVLDEDA PPEKRAGVHD GHLKRAPKVY CGGDERDVLR VGSGGFWPRR SRLWGGVDHA PAGFNPTVTV FHVYDILENV EHAYGMRAAQ FHARFMDAIT PTGTVITLLG LTPEGHRVAV HVYGTRQYFY ...String: HHHHHHNFYN PYLAPVGTQQ KPTGPTQRHT YYSECDEFRF IAPRVLDEDA PPEKRAGVHD GHLKRAPKVY CGGDERDVLR VGSGGFWPRR SRLWGGVDHA PAGFNPTVTV FHVYDILENV EHAYGMRAAQ FHARFMDAIT PTGTVITLLG LTPEGHRVAV HVYGTRQYFY MNKEEVDRHL QCRAPRDLCE RMAAALRESP GASFRGISAD HFEAEVVERT DVYYYETRPA LFYRVYVRSG RVLSYLCDNF CPAIKKYEGG VDATTRFILD NPGFVTFGWY RLKPGRNNTL AQPRAPMAFG TSSDVEFNCT ADNLAIEGGM SDLPAYKLMC FDIECKAGGE DELAFPVAGH PEDLVIQISC LLYDLSTTAL EHVLLFSLGS CDLPESHLNE LAARGLPTPV VLEFDSEFEM LLAFMTLVKQ YGPEFVTGYN IINFDWPFLL AKLTDIYKVP LDGYGRMNGR GVFRVWDIGQ SHFQKRSKIK VNGMVNIDMY GIITDKIKLS SYKLNAVAEA VLKDKKKDLS YRDIPAYYAT GPAQRGVIGE YCIQDSLLVG QLFFKFLPHL ELSAVARLAG INITRTIYDG QQIRVFTCLL RLADQKGFIL PDTQGRFRGA GGEAPKRPAA AREDEERPEE EGEDEDEREE GGGEREPEGA RETAGRHVGY QGARVLDPTS GFHVNPVVVF DFASLYPSII QAHNLCFSTL SLRADAVAHL EAGKDYLEIE VGGRRLFFVK AHVRESLLSI LLRDVLAMRK QIRSRIPQSS PEEAVLLDKQ QAAIKVVCNS VYGFTGVQHG LLPCLHVAAT VTTIGREMLL ATREYVHARW AAFEQLLADF PEAADMRAPG PYSMRIIYGD TDSIFVLCRG LTAAGLTAMG DKMASHISRA LFLPPIKLEC EKTFTKLLLI AKKKYIGVIY GGKMLIKGVD LVRKNNCAFI NRTSRALVDL LFYDDTVSGA AAALAERPAE EWLARPLPEG LQAFGAVLVD AHRRITDPER DIQDFVLTAE LSRHPRAYTN KRLAHLTVYY KLMARRAQVP SIKDRIPYVI VAQTREVEET VARLAALREL DAAAPGDEPA PPAALPSPAK RPRETPSHAD PPGGASKPRK LLVSELAEDP AYAIAHGVAL NTDYYFSHLL GAACVTFKAL FGNNAKITES LLKRFIPEVW HPPDDVAARL RAAGFGAVGA GATAEETRRM LHRAFDTLA UniProtKB: UNIPROTKB: AFE62858 |
-Macromolecule #2: DNA polymerase processivity factor
| Macromolecule | Name: DNA polymerase processivity factor / type: protein_or_peptide / ID: 2 Details: Herpes simplex virus type 1 (KOS strain) processivity factor UL42 residues 1-340 preceding a maltose binding protein (MBP) tag and a PreScission protease cleavage site (MBP-PP-UL42dC340)) Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS Human alphaherpesvirus 1 strain KOS |
| Recombinant expression | Organism:  |
| Sequence | String: MTDSPGGVAP ASPVEDASDA SLGQPEEGAP CQVVLQGAEL NGILQAFAPL RTSLLDSLLV MGDRGILIHN TIFGEQVFLP LEHSQFSRYR WRGPTAAFLS LVDQKRSLLS VFRANQYPDL RRVELAITGQ APFRTLVQRI WTTTSDGEAV ELASETLMKR ELTSFVVLVP ...String: MTDSPGGVAP ASPVEDASDA SLGQPEEGAP CQVVLQGAEL NGILQAFAPL RTSLLDSLLV MGDRGILIHN TIFGEQVFLP LEHSQFSRYR WRGPTAAFLS LVDQKRSLLS VFRANQYPDL RRVELAITGQ APFRTLVQRI WTTTSDGEAV ELASETLMKR ELTSFVVLVP QGTPDVQLRL TRPQLTKVLN ATGADSATPT TFELGVNGKF SVFTTSTCVT FAAREEGVSS STSTQVQILS NALTKAGQAA ANAKTVYGEN THRTFSVVVD DCSMRAVLRR LQVAGGTLKF FLTTPVPSLC VTATGPNAVS AVFLLKPQKI CLDWLGHSQG SPSAGSSASR UniProtKB: UNIPROTKB: AFE62870 |
-Macromolecule #3: DNA primer
| Macromolecule | Name: DNA primer / type: dna / ID: 3 / Details: 3'-deoxy primer DNA strand / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: GATTACGAAT TCGAGCTCGG TACCCGGGGA T(DOC) |
-Macromolecule #4: DNA template
| Macromolecule | Name: DNA template / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: CACACACACA CACACACAGA TCCCCGGGTA CCGAGCTCGA ATTCGTAATC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.38 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM tris(2-carboxyethyl)phosphine (TCEP) |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Pelco easiGlow at 15 mA for 30 seconds under 0.39 mBar (i.e. 39 Pa) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 3 microliters of sample were blotted for 3 seconds with filter paper saturated under 100% humidity prior to plunging.. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 1678 / Average exposure time: 1.5 sec. / Average electron dose: 58.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 60606 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)