[English] 日本語
 Yorodumi
Yorodumi- EMDB-42855: Cryo-EM structure of the unliganded hexameric prenyltransferase i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the unliganded hexameric prenyltransferase in bifunctional copalyl diphosphate synthase from Penicillium fellutanum with an open conformation, reconstruction in C1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | terpene / biosynthesis / enzyme / TRANSFERASE | |||||||||
| Biological species |  Penicillium fellutanum ATCC 48694 (fungus) Penicillium fellutanum ATCC 48694 (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Gaynes MN / Christianson DW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2024 Journal: J Struct Biol / Year: 2024Title: Structure of the prenyltransferase in bifunctional copalyl diphosphate synthase from Penicillium fellutanum reveals an open hexamer conformation. Authors: Matthew N Gaynes / Trey A Ronnebaum / Kollin Schultz / Jacque L Faylo / Ronen Marmorstein / David W Christianson /  Abstract: Copalyl diphosphate synthase from Penicillium fellutanum (PfCPS) is an assembly-line terpene synthase that contains both prenyltransferase and class II cyclase activities. The prenyltransferase ...Copalyl diphosphate synthase from Penicillium fellutanum (PfCPS) is an assembly-line terpene synthase that contains both prenyltransferase and class II cyclase activities. The prenyltransferase catalyzes processive chain elongation reactions using dimethylallyl diphosphate and three equivalents of isopentenyl diphosphate to yield geranylgeranyl diphosphate, which is then utilized as a substrate by the class II cyclase domain to generate copalyl diphosphate. Here, we report the 2.81 Å-resolution cryo-EM structure of the hexameric prenyltransferase of full-length PfCPS, which is surrounded by randomly splayed-out class II cyclase domains connected by disordered polypeptide linkers. The hexamer can be described as a trimer of dimers; surprisingly, one of the three dimer-dimer interfaces is separated to yield an open hexamer conformation, thus breaking the D3 symmetry typically observed in crystal structures of other prenyltransferase hexamers such as wild-type human GGPP synthase (hGGPPS). Interestingly, however, an open hexamer conformation was previously observed in the crystal structure of D188Y hGGPPS, apparently facilitated by hexamer-hexamer packing in the crystal lattice. The cryo-EM structure of the PfCPS prenyltransferase hexamer is the first to reveal that an open conformation can be achieved even in the absence of a point mutation or interaction with another hexamer. Even though PfCPS octamers are not detected, we suggest that the open hexamer conformation represents an intermediate in the hexamer-octamer equilibrium for those prenyltransferases that do exhibit oligomeric heterogeneity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42855.map.gz emd_42855.map.gz | 643.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42855-v30.xml emd-42855-v30.xml emd-42855.xml emd-42855.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42855_fsc.xml emd_42855_fsc.xml | 19.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_42855.png emd_42855.png | 51.9 KB | ||
| Filedesc metadata |  emd-42855.cif.gz emd-42855.cif.gz | 5.6 KB | ||
| Others |  emd_42855_additional_1.map.gz emd_42855_additional_1.map.gz emd_42855_half_map_1.map.gz emd_42855_half_map_1.map.gz emd_42855_half_map_2.map.gz emd_42855_half_map_2.map.gz | 361.7 MB 676 MB 676 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42855 http://ftp.pdbj.org/pub/emdb/structures/EMD-42855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42855 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42855.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42855.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.55 Å | ||||||||||||||||||||||||||||||||||||
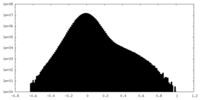
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened C1 map of the hexameric PfCPS prenyltransferase...
| File | emd_42855_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened C1 map of the hexameric PfCPS prenyltransferase in non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
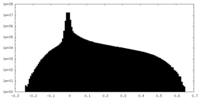
| Density Histograms |
-Half map: #1
| File | emd_42855_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
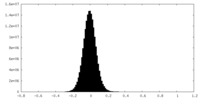
| Density Histograms |
-Half map: #2
| File | emd_42855_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
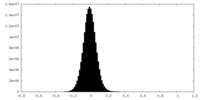
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric prenyltransferase from the bifunctional copalyl diphosp...
| Entire | Name: Hexameric prenyltransferase from the bifunctional copalyl diphosphate synthase of Penicillium fellutanum |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric prenyltransferase from the bifunctional copalyl diphosp...
| Supramolecule | Name: Hexameric prenyltransferase from the bifunctional copalyl diphosphate synthase of Penicillium fellutanum type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Penicillium fellutanum ATCC 48694 (fungus) Penicillium fellutanum ATCC 48694 (fungus) |
| Molecular weight | Theoretical: 634.002 KDa |
-Macromolecule #1: Copalyl diphosphate synthase
| Macromolecule | Name: Copalyl diphosphate synthase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Penicillium fellutanum ATCC 48694 (fungus) Penicillium fellutanum ATCC 48694 (fungus) |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGENLYFQG HMASMESASL DQSAALLVKE LTEHIDDSNG LGFMSPAIYD TAWVSMIKKT DNDQTFWLFP KSFHYILENQ LENGGWVTYA SEIDGILNTS ASLLSLKRHF DLPLQISTES QTSMENRIRK ATDALRVLLR TWDVDATLHV GFEILVPALL ...String: MGSSHHHHHH SSGENLYFQG HMASMESASL DQSAALLVKE LTEHIDDSNG LGFMSPAIYD TAWVSMIKKT DNDQTFWLFP KSFHYILENQ LENGGWVTYA SEIDGILNTS ASLLSLKRHF DLPLQISTES QTSMENRIRK ATDALRVLLR TWDVDATLHV GFEILVPALL DYLQVEGLTF DFPGRDKLFQ IRDQKLSRFK PEFIYAPFQT TALHSLEAFI GLIDFDRVQH HKVRGSFMAS PSSTAAVLMN ATEWDIDCEE YIRHVIEHAS GKASGGVPSA FPSTIFEITW TLSTLLKAGF NLSSNDSSNV QKACSYLLGV LTAEKGAIGF VPSVCADADD TAKTILVLSL LRENVLPDGM LKAFEVENHF KTYPLERDPS FSANCNVLLA LLHLENPSQY TLQIEKATRF LYTHFRESNL NVRDKWNLSP FYSWMLMAQA IARLDELCKG SQLKNLHDHV ESDLIPLLQE MTVSVMHQQN KDGSWGTKLS KEETAYAVLM LTYAVSFEAL VGPRRQIRNA IEEGCLFLRS GKDATSERLW VEKVTYESQM LSRAYTLAAL KNALDVLKKD DMKIAFIGNS VNESFTEIEV VNGKNVTEPS SNTYDLKEVK MDKAEFTPPD TPPRSKSTSV DPIEEAICAH ENRGAISDHT SRAIDLCRNP PLWGTDQEQT LLGPFEYLES IPGKNIRSQF IEAFNTWLQI PQDHLQIVGK VISMLHTASL LVDDIEDNSL LRRGQPVAHS IFGTAQTFNS GNYVYFLALQ EVQKLNSPRA ISIFVDALTQ LHRGQGMDVF WRDSLICPTE EEYLDMVANK TGALFCLAIE LLQIKSTVQL DFLPLVRLLG IIFQICDDYL NLKSTNYTQK KGLCEDITEG KFSFPIIHSI RTKPGNRQLI NVLRQKSKED DVKRFALAYM ESTQSFDYTR DFVKILNGEA LRMIEDLEQQ GLHRNIEIRN ILARMSLEQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 12467 / Average exposure time: 2.15 sec. / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)