[English] 日本語
 Yorodumi
Yorodumi- EMDB-42514: Structural and biochemical investigations of a HEAT-repeat protei... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | IRON-SULFUR CLUSTER / METALLOCOFACTOR / ASSEMBLY / METAL TRANSPORT | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.43 Å | |||||||||||||||
 Authors Authors | Vasquez S / Drennan CL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway. Authors: Sheena Vasquez / Melissa D Marquez / Edward J Brignole / Amanda Vo / Sunnie Kong / Christopher Park / Deborah L Perlstein / Catherine L Drennan /  Abstract: Iron-sulfur clusters are essential for life and defects in their biosynthesis lead to human diseases. The mechanism of cluster assembly and delivery to cytosolic and nuclear client proteins via the ...Iron-sulfur clusters are essential for life and defects in their biosynthesis lead to human diseases. The mechanism of cluster assembly and delivery to cytosolic and nuclear client proteins via the cytosolic iron-sulfur cluster assembly (CIA) pathway is not well understood. Here we report cryo-EM structures of the HEAT-repeat protein Met18 from Saccharomyces cerevisiae, a key component of the CIA targeting complex (CTC) that identifies cytosolic and nuclear client proteins and delivers a mature iron-sulfur cluster. We find that in the absence of other CTC proteins, Met18 adopts tetrameric and hexameric states. Using mass photometry and negative stain EM, we show that upon the addition of Cia2, these higher order oligomeric states of Met18 disassemble. We also use pulldown assays to identify residues of critical importance for Cia2 binding and recognition of the Leu1 client, many of which are buried when Met18 oligomerizes. Our structures show conformations of Met18 that have not been previously observed in any Met18 homolog, lending support to the idea that a highly flexible Met18 may be key to how the CTC is able to deliver iron-sulfur clusters to client proteins of various sizes and shapes, i.e. Met18 conforms to the dimensions needed. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42514.map.gz emd_42514.map.gz | 70.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42514-v30.xml emd-42514-v30.xml emd-42514.xml emd-42514.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42514.png emd_42514.png | 78.8 KB | ||
| Filedesc metadata |  emd-42514.cif.gz emd-42514.cif.gz | 5.8 KB | ||
| Others |  emd_42514_half_map_1.map.gz emd_42514_half_map_1.map.gz emd_42514_half_map_2.map.gz emd_42514_half_map_2.map.gz | 69.3 MB 69.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42514 http://ftp.pdbj.org/pub/emdb/structures/EMD-42514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42514 | HTTPS FTP |
-Validation report
| Summary document |  emd_42514_validation.pdf.gz emd_42514_validation.pdf.gz | 787.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42514_full_validation.pdf.gz emd_42514_full_validation.pdf.gz | 787.5 KB | Display | |
| Data in XML |  emd_42514_validation.xml.gz emd_42514_validation.xml.gz | 12.9 KB | Display | |
| Data in CIF |  emd_42514_validation.cif.gz emd_42514_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42514 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42514 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42514.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42514.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5998 Å | ||||||||||||||||||||||||||||||||||||
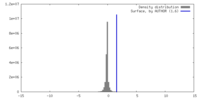
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42514_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
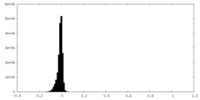
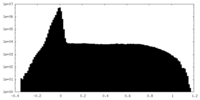
| Density Histograms |
-Half map: #1
| File | emd_42514_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
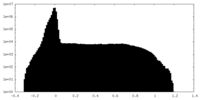
| Density Histograms |
- Sample components
Sample components
-Entire : QUATERNARY COMPLEX OF MET18 TETRAMER
| Entire | Name: QUATERNARY COMPLEX OF MET18 TETRAMER |
|---|---|
| Components |
|
-Supramolecule #1: QUATERNARY COMPLEX OF MET18 TETRAMER
| Supramolecule | Name: QUATERNARY COMPLEX OF MET18 TETRAMER / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: MET18 HEXAMER IMAGED AND SOLVED BY CRYO-EM. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 118 kDa/nm |
-Macromolecule #1: Met18/MMS19
| Macromolecule | Name: Met18/MMS19 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: AVVTFMANLN IDDSKANETA STVTDSIVHR SIKLLEVVVA LKDYFLSENE VERKKALTCL TTILAKTPKD HLSKNECSV IFQFYQSKLD DQALAKEVLE GFAALAPMKY VSINEIAQLL RLLLDNYQQG QHLASTRLWP F KILRKIFD RFFVNGSSTE QVKRINDLFI ...String: AVVTFMANLN IDDSKANETA STVTDSIVHR SIKLLEVVVA LKDYFLSENE VERKKALTCL TTILAKTPKD HLSKNECSV IFQFYQSKLD DQALAKEVLE GFAALAPMKY VSINEIAQLL RLLLDNYQQG QHLASTRLWP F KILRKIFD RFFVNGSSTE QVKRINDLFI ETFLHVANGE KDPRNLLLSF ALNKSITSSL QNVENAKEDL FD VLFCYFA ALKTALRSAI TATPLFAEDA YSNLLDKLTA SSPVVKNDTL LTLLECVRKF GGSSILENWT LLW NALKFE IMQNYTNYDA CLKIINLMAL QLYNFDKVSF EKFFTHVLDE LKPNFKYEKD LKQTCQILSA IGSG NVEIF NKVISSTFPL FLINTSEVAK LKLLIMNFSF FVDSYIDLFG RTSKESLGTP VPNNKMAEYK DEIIM ILSM ALTRSSKAEV TIRTLSVIQF TKMIKMKGFL TPEEVSLIIQ YFTEEILTDN NKNIYYACLE GLKTIS EIY EDLVFEISLK KLLDLLPDCF EEKIRVNDEE NIHIETILKI ILDFTTSRHI LVKESITFLA TKLNRVA KI SKSREYCFLL ISTIYSLFNN NNQNENVLNE EDALALKNAI EPKLFEIITQ ESAIVSDNYN LTLLSNVL F FTNLKIPQAA HQEELDRYNE LFISEGKIRI LDTPNVLAIS YAKILSALNK NCQFPQKFTV LFGTVQLLK KHAPRMTETE KLGYLELLLV LSNKFVSEKD VIGLFDWKDL SVINLEVMVW LTKGLIMQNS LESSEIAKKF IDLLSNEEI GSLVSKLFEV FVMDISSLKK FKGISWNNNV KILYKQKFFG DIFQTLVSNY KNTVDMTIKC N YLTALSLV LKHTPSQSVG PFINDLFPLL LQALDMPDPE VRVSALETLK DTTDKHHTLI TEHVSTIVPL LL SLSLPHK YNSVSVRLIA LQLLEMITTV VPLNYCLSYQ DDVLSALIPV LSDKKRIIRK QCVDTRQVYY ELG QI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 77 % / Chamber temperature: 297.15 K / Details: SAMPLE WAS PREPARED ON THE CHAMELEON. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Software | Name: EPU |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 53.47 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 92000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.3 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Software | Name: PHENIX |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)