[English] 日本語
 Yorodumi
Yorodumi- EMDB-42250: Varicella-zoster virus glycoprotein B; H527P prefusion class I. -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Varicella-zoster virus glycoprotein B; H527P prefusion class I. | ||||||||||||||||||||||||||||||
 Map data Map data | Cryo-ET map of the VZV gB mutant H527P; class I | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Herpesvirus / Fusogen / Glycoprotein B / Varicella Zoster-Virus / VIRAL PROTEIN | ||||||||||||||||||||||||||||||
| Biological species |  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) | ||||||||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 9.8 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Zhou M / Oliver SL / Muyuan C / Vollmer B | ||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  Germany, European Union, 9 items Germany, European Union, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Targeted mutagenesis of the herpesvirus fusogen central helix captures transition states. Authors: Momei Zhou / Benjamin Vollmer / Emily Machala / Muyuan Chen / Kay Grünewald / Ann M Arvin / Wah Chiu / Stefan L Oliver /   Abstract: Herpesviruses remain a burden for animal and human health, including the medically important varicella-zoster virus (VZV). Membrane fusion mediated by conserved core glycoproteins, the fusogen gB and ...Herpesviruses remain a burden for animal and human health, including the medically important varicella-zoster virus (VZV). Membrane fusion mediated by conserved core glycoproteins, the fusogen gB and the heterodimer gH-gL, enables herpesvirus cell entry. The ectodomain of gB orthologs has five domains and is proposed to transition from a prefusion to postfusion conformation but the functional relevance of the domains for this transition remains poorly defined. Here we describe structure-function studies of the VZV gB DIII central helix targeting residues EHV. Critically, a H527P mutation captures gB in a prefusion conformation as determined by cryo-EM, a loss of membrane fusion in a virus free assay, and failure of recombinant VZV to spread in cell monolayers. Importantly, two predominant cryo-EM structures of gB[H527P] are identified by 3D classification and focused refinement, suggesting they represented gB conformations in transition. These studies reveal gB DIII as a critical element for herpesvirus gB fusion function. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42250.map.gz emd_42250.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42250-v30.xml emd-42250-v30.xml emd-42250.xml emd-42250.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42250.png emd_42250.png | 62.9 KB | ||
| Filedesc metadata |  emd-42250.cif.gz emd-42250.cif.gz | 5.6 KB | ||
| Others |  emd_42250_half_map_1.map.gz emd_42250_half_map_1.map.gz emd_42250_half_map_2.map.gz emd_42250_half_map_2.map.gz | 16.8 MB 16.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42250 http://ftp.pdbj.org/pub/emdb/structures/EMD-42250 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42250 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42250 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42250.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42250.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-ET map of the VZV gB mutant H527P; class I | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.19 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map even
| File | emd_42250_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map even | ||||||||||||
| Projections & Slices |
| ||||||||||||
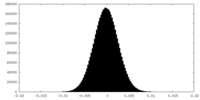
| Density Histograms |
-Half map: Half map odd
| File | emd_42250_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map odd | ||||||||||||
| Projections & Slices |
| ||||||||||||
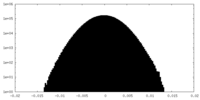
| Density Histograms |
- Sample components
Sample components
-Entire : Human alphaherpesvirus 3
| Entire | Name:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
|---|---|
| Components |
|
-Supramolecule #1: Human alphaherpesvirus 3
| Supramolecule | Name: Human alphaherpesvirus 3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Extracellular vesicles produced by BHK-21 cells transfected with a plasmid expressing VZV glycoprotein B mutant H527P. NCBI-ID: 10335 / Sci species name: Human alphaherpesvirus 3 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Varicella-zoster virus gB[H527P] mutant
| Macromolecule | Name: Varicella-zoster virus gB[H527P] mutant / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: MSPCGYYSKW RNRDRPEYRR NLRFRRFFSS IHPNAAAGSG FNGPGVFITS VTGVWLCFLC IFSMFVTAVV SVSPSSFYES LQVEPTQSED ITRSAHLGDG DEIREAIHKS QDAETKPTFY VCPPPTGSTI VRLEPPRTCP DYHLGKNFTE GIAVVYKENI AAYKFKATVY ...String: MSPCGYYSKW RNRDRPEYRR NLRFRRFFSS IHPNAAAGSG FNGPGVFITS VTGVWLCFLC IFSMFVTAVV SVSPSSFYES LQVEPTQSED ITRSAHLGDG DEIREAIHKS QDAETKPTFY VCPPPTGSTI VRLEPPRTCP DYHLGKNFTE GIAVVYKENI AAYKFKATVY YKDVIVSTAW AGSSYTQITN RYADRVPIPV SEITDTIDKF GKCSSKATYV RNNHKVEAFN EDKNPQDMPL IASKYNSVGS KAWHTTNDTY MVAGTPGTYR TGTSVNCIIE EVEARSIFPY DSFGLSTGDI IYMSPFFGLR DGAYREHSNY AMDRFHQFEG YRQRDLDTRA LLEPAARNFL VTPHLTVGWN WKPKRTEVCS LVKWREVEDV VRDEYAHNFR FTMKTLSTTF ISETNEFNLN QIHLSQCVKE EARAIINRIY TTRYNSSHVR TGDIQTYLAR GGFVVVFQPL LSNSLARLYL QELVRENTNH SPQKHPTRNT RSRRSVPVEL RANRTITTTS SVEFAMLQFT YDHIQEPVNE MLARISSSWC QLQNRERALW SGLFPINPSA LASTILDQRV KARILGDVIS VSNCPELGSD TRIILQNSMR VSGSTTRCYS RPLISIVSLN GSGTVEGQLG TDNELIMSRD LLEPCVANHK RYFLFGHHYV YYEDYRYVRE IAVHDVGMIS TYVDLNLTLL KDREFMPLRV YTRDELRDTG LLDYSEIQRR NQMHSLRFYD IDKVVQYDSG TAIMQGMAQF FQGLGTAGQA VGHVVLGATG ALLSTVHGFT TFLSNPFGAL AVGLLVLAGL VAAFFAYRYV LKLKTSPMKA LYPLTTKGLK QLPEGMDPFA EKPNATDTPI EEIGDSQNTE PSVNSGFDPD KFREAQEMIK YMTLVSAAER QESKARKKNK TSALLTSRLT GLALRNRRGY SRVRTENVTG V |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 0.3 sec. / Average electron dose: 3.22 e/Å2 / Details: A total of 43 tilt series. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.002 µm / Nominal defocus min: 0.002 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 9.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN2 / Number subtomograms used: 43 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 2074 |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)