[English] 日本語
 Yorodumi
Yorodumi- EMDB-42164: The CryoEM structure of the high affinity Carbon monoxide dehydro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The CryoEM structure of the high affinity Carbon monoxide dehydrogenase from Mycobacterium smegmatis | |||||||||
 Map data Map data | Primary Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Carbon monoxide dehydrogenase / MoCu / Mycobacterium smegmatis / High affinity / trace gas scavenging / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbon-monoxide dehydrogenase (acceptor) / anaerobic carbon-monoxide dehydrogenase activity / molybdenum ion binding / FAD binding / 2 iron, 2 sulfur cluster binding / oxidoreductase activity / iron ion binding / copper ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.85 Å | |||||||||
 Authors Authors | Grinter R / Venugopal H / Greening C / Gillett D | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2025 Journal: Nat Chem Biol / Year: 2025Title: Quinone extraction drives atmospheric carbon monoxide oxidation in bacteria. Authors: Ashleigh Kropp / David L Gillett / Hari Venugopal / Miguel A Gonzálvez / James P Lingford / Surbhi Jain / Christopher K Barlow / Jie Zhang / Chris Greening / Rhys Grinter /   Abstract: Diverse bacteria and archaea use atmospheric CO as an energy source for long-term survival. Bacteria use [MoCu]-CO dehydrogenases (Mo-CODH) to convert atmospheric CO to carbon dioxide, transferring ...Diverse bacteria and archaea use atmospheric CO as an energy source for long-term survival. Bacteria use [MoCu]-CO dehydrogenases (Mo-CODH) to convert atmospheric CO to carbon dioxide, transferring the obtained electrons to the aerobic respiratory chain. However, it is unknown how these enzymes oxidize CO at low concentrations and interact with the respiratory chain. Here, we use cryo-electron microscopy and structural modeling to show how Mo-CODH (CoxSML) from Mycobacterium smegmatis interacts with its partner, the membrane-bound menaquinone-binding protein CoxG. We provide electrochemical, biochemical and genetic evidence that Mo-CODH transfers CO-derived electrons to the aerobic respiratory chain through CoxG. Lastly, we show that Mo-CODH and CoxG genetically and structurally associate in diverse bacteria and archaea. These findings reveal the basis of the biogeochemically and ecologically important process of atmospheric CO oxidation, while demonstrating that long-range quinone transport is a general mechanism of energy conservation, which convergently evolved on multiple occasions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42164.map.gz emd_42164.map.gz | 122.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42164-v30.xml emd-42164-v30.xml emd-42164.xml emd-42164.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42164_fsc.xml emd_42164_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42164.png emd_42164.png | 95 KB | ||
| Filedesc metadata |  emd-42164.cif.gz emd-42164.cif.gz | 7.2 KB | ||
| Others |  emd_42164_half_map_1.map.gz emd_42164_half_map_1.map.gz emd_42164_half_map_2.map.gz emd_42164_half_map_2.map.gz | 120.4 MB 120.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42164 http://ftp.pdbj.org/pub/emdb/structures/EMD-42164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42164 | HTTPS FTP |
-Validation report
| Summary document |  emd_42164_validation.pdf.gz emd_42164_validation.pdf.gz | 997.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42164_full_validation.pdf.gz emd_42164_full_validation.pdf.gz | 997.1 KB | Display | |
| Data in XML |  emd_42164_validation.xml.gz emd_42164_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_42164_validation.cif.gz emd_42164_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42164 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42164 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42164 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42164 | HTTPS FTP |
-Related structure data
| Related structure data |  8uemMC  8udsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42164.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42164.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_42164_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_42164_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MoCu Carbon monoxide Dehydrogenase (MoCu-CODH)
| Entire | Name: MoCu Carbon monoxide Dehydrogenase (MoCu-CODH) |
|---|---|
| Components |
|
-Supramolecule #1: MoCu Carbon monoxide Dehydrogenase (MoCu-CODH)
| Supramolecule | Name: MoCu Carbon monoxide Dehydrogenase (MoCu-CODH) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 270 KDa |
-Macromolecule #1: Carbon monoxide dehydrogenase (Large chain), CoxL
| Macromolecule | Name: Carbon monoxide dehydrogenase (Large chain), CoxL / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 85.957844 KDa |
| Sequence | String: MTTLENPVER PEDTAVNDAK PCGHGRMLRK EDPRFIRGRG NYVDDVKLPG MLHLAILRSP YAHATINSID VTAAQAHPKV KAVVTGADL AAKGLAWMPT LSNDVQAVLA TDKVRFQGQE VAFVVAEDRY SARDALELID VDYEPLDPVI DARHALDPGA P VIRTDLDG ...String: MTTLENPVER PEDTAVNDAK PCGHGRMLRK EDPRFIRGRG NYVDDVKLPG MLHLAILRSP YAHATINSID VTAAQAHPKV KAVVTGADL AAKGLAWMPT LSNDVQAVLA TDKVRFQGQE VAFVVAEDRY SARDALELID VDYEPLDPVI DARHALDPGA P VIRTDLDG KTDNHCFDWE TGDAAATDAV FAKADVVVKQ EMVYPRVHPA PMETCGAVAD LDPVTRKLTL WSTTQAPHAH RT LYALVAG LPEHKIRVIS PDIGGGFGNK VPIYPGYVCA IVGSLLLGKP VKWMEDRSEN LTSTGFARDY IMVGEIAATR DGK ILAIRS NVLADHGAFN GTAAPVKYPA GFFGVFTGSY DIEAAYCHMT AVYTNKAPGG VAYACSFRIT EAVYFVERLV DCLA YELKM DPAQLRLQNL LKAEQFPYTS KTGWVYDSGD YEKTMRLAME MVDYEGLRAE QAEKRKRGEL MGIGMSFFTE AVGAG PRKD MDILGLGMAD GCELRVHPTG KAVVRLSVQS QGQGHETTFA QIVAEELGIP PEDIDVVHGD TDQTPFGLGT YGSRST PVS GAAAALVARK VRDKAKIIAA GMLEASIADL EWDKGSFHIK GDPSASVTIA DIAMRAHGAG DLPEGLEGGL DAQICYN PS NLTYPYGAYF CVVDIDPGTA VVKVRRFVAV DDCGTRINPM IIEGQIHGGL VDGIGMALME MIAFDEDGNC LGGSLMDY L IPTAMEVPHF ETGHTVTPSP HHPIGAKGIG ESATVGSPPA VVNAVVDALA PYGVRHADMP LTPSRVWEAM QGRATPPI UniProtKB: Carbon monoxide dehydrogenase (Large chain), CoxL |
-Macromolecule #2: Carbon monoxide dehydrogenase medium chain
| Macromolecule | Name: Carbon monoxide dehydrogenase medium chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 31.710051 KDa |
| Sequence | String: MQVPGPFEYE RATSVDHAVG LLDRLGEDAR IVAGGHSLLP MMKLRIANPE YLVDINDLAV ELGYVITDPT LVRIGAMARH RQVLESDPL AAVCPIFRDA ERVIADPVVR NRGTLGGSLC QADPAEDLTT VCTILGAVCL ARGPGGEREI GIDDFLVGPY E TALAHNEM ...String: MQVPGPFEYE RATSVDHAVG LLDRLGEDAR IVAGGHSLLP MMKLRIANPE YLVDINDLAV ELGYVITDPT LVRIGAMARH RQVLESDPL AAVCPIFRDA ERVIADPVVR NRGTLGGSLC QADPAEDLTT VCTILGAVCL ARGPGGEREI GIDDFLVGPY E TALAHNEM LVEVRIPVRH RTSSAYAKVE RRVGDWAVTA AGAQVTLDGD SIVAARVGLT AVNPDPDALR ALADDLIGKP AT EETFAAA GELAVQACEP VTDTRGSADY KRHLARELTI RTMRTAVERV RTAPAPEGN UniProtKB: Carbon monoxide dehydrogenase medium chain |
-Macromolecule #3: [2Fe-2S] binding domain protein
| Macromolecule | Name: [2Fe-2S] binding domain protein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 17.14857 KDa |
| Sequence | String: MQVTMTVNGE AVTADVEPRM LLVHFLRDQL GLTGTHWGCD TSNCGTCVVE VDGEPVKSCT MLAAMASGHS VNTVEGMEVD GKLDPVQEG FMQCHGLQCG FCTPGMMITA RALLRQNPDP TEEEIREAIS GQICRCTGYT TIVRSVQWAA RHAREEAKA UniProtKB: [2Fe-2S] binding domain protein |
-Macromolecule #4: CU(I)-S-MO(IV)(=O)OH CLUSTER
| Macromolecule | Name: CU(I)-S-MO(IV)(=O)OH CLUSTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: CUN |
|---|---|
| Molecular weight | Theoretical: 224.558 Da |
| Chemical component information | 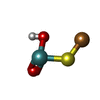 ChemComp-CUN: |
-Macromolecule #5: PTERIN CYTOSINE DINUCLEOTIDE
| Macromolecule | Name: PTERIN CYTOSINE DINUCLEOTIDE / type: ligand / ID: 5 / Number of copies: 2 / Formula: MCN |
|---|---|
| Molecular weight | Theoretical: 696.501 Da |
| Chemical component information |  ChemComp-MCN: |
-Macromolecule #6: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 6 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #7: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 7 / Number of copies: 4 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 899 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 / Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/4 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.001 kPa | |||||||||
| Vitrification | Cryogen name: PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4508 / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8uem: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































