+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the flagellar MotAB stator bound to FliG | |||||||||
 Map data Map data | Global B-factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | flagella chemotaxis motility type III secretion system / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum basal body / bacterial-type flagellum-dependent swarming motility / cytoskeletal motor activity / bacterial-type flagellum-dependent cell motility / proton transmembrane transport / chemotaxis / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Clostridium sporogenes (bacteria) Clostridium sporogenes (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Deme JC / Johnson S / Lea SM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Structural basis of directional switching by the bacterial flagellum. Authors: Steven Johnson / Justin C Deme / Emily J Furlong / Joseph J E Caesar / Fabienne F V Chevance / Kelly T Hughes / Susan M Lea /    Abstract: The bacterial flagellum is a macromolecular protein complex that harvests energy from uni-directional ion flow across the inner membrane to power bacterial swimming via rotation of the flagellar ...The bacterial flagellum is a macromolecular protein complex that harvests energy from uni-directional ion flow across the inner membrane to power bacterial swimming via rotation of the flagellar filament. Rotation is bi-directional, with binding of a cytoplasmic chemotactic response regulator controlling reversal, though the structural and mechanistic bases for rotational switching are not well understood. Here we present cryoelectron microscopy structures of intact Salmonella flagellar basal bodies (3.2-5.5 Å), including the cytoplasmic C-ring complexes required for power transmission, in both counter-clockwise and clockwise rotational conformations. These reveal 180° movements of both the N- and C-terminal domains of the FliG protein, which, when combined with a high-resolution cryoelectron microscopy structure of the MotAB stator, show that the stator shifts from the outside to the inside of the C-ring. This enables rotational switching and reveals how uni-directional ion flow across the inner membrane is used to accomplish bi-directional rotation of the flagellum. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42139.map.gz emd_42139.map.gz | 323.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42139-v30.xml emd-42139-v30.xml emd-42139.xml emd-42139.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42139_fsc.xml emd_42139_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42139.png emd_42139.png | 159.6 KB | ||
| Masks |  emd_42139_msk_1.map emd_42139_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42139.cif.gz emd-42139.cif.gz | 6.7 KB | ||
| Others |  emd_42139_additional_1.map.gz emd_42139_additional_1.map.gz emd_42139_additional_2.map.gz emd_42139_additional_2.map.gz emd_42139_half_map_1.map.gz emd_42139_half_map_1.map.gz emd_42139_half_map_2.map.gz emd_42139_half_map_2.map.gz | 172.4 MB 283.6 MB 318.5 MB 318.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42139 http://ftp.pdbj.org/pub/emdb/structures/EMD-42139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42139 | HTTPS FTP |
-Related structure data
| Related structure data |  8ucsMC  8umdC  8umxC  8uoxC  8uplC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42139.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42139.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global B-factor sharpened map | ||||||||||||||||||||||||||||||||||||
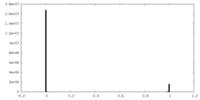
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.693 Å | ||||||||||||||||||||||||||||||||||||
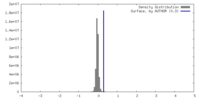
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42139_msk_1.map emd_42139_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
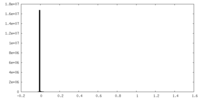
| Projections & Slices |
| ||||||||||||
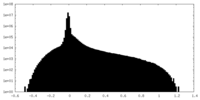
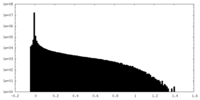
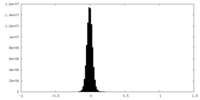
| Density Histograms |
-Additional map: unsharpened map
| File | emd_42139_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened with deepEMhancer
| File | emd_42139_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened with deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_42139_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_42139_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MotAB stator complexed with FliG
| Entire | Name: MotAB stator complexed with FliG |
|---|---|
| Components |
|
-Supramolecule #1: MotAB stator complexed with FliG
| Supramolecule | Name: MotAB stator complexed with FliG / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Clostridium sporogenes (bacteria) Clostridium sporogenes (bacteria) |
-Macromolecule #1: OmpA family protein
| Macromolecule | Name: OmpA family protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium sporogenes (bacteria) Clostridium sporogenes (bacteria) |
| Molecular weight | Theoretical: 32.078861 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARRNKKGGG GDEIRGDEWL ATFSDTITLL LTFFILLYSF SSVDAQKFQQ VASAMQVAMT GQSGNTIVDY NLKNGDVPLV GETTKLGRE TGSDAKSDSK EVYNEVNKFV DKNNLKSSVE VKEDGRGVII QLRDNVLFEI GRADIKPQSK QIMDKINGLI A TLPNEVIV ...String: MARRNKKGGG GDEIRGDEWL ATFSDTITLL LTFFILLYSF SSVDAQKFQQ VASAMQVAMT GQSGNTIVDY NLKNGDVPLV GETTKLGRE TGSDAKSDSK EVYNEVNKFV DKNNLKSSVE VKEDGRGVII QLRDNVLFEI GRADIKPQSK QIMDKINGLI A TLPNEVIV EGHTDNVPIK NEVYGSNWEL STARAVNVLR YFVETKKQNP VRFTAAGYGE YRPIAQNNSD VNKAKNRRVN IV IVSKEKE SSKKENLYFQ GQFGSWSHPQ FEKGGGSGGG SGGGSWSHPQ FEK UniProtKB: OmpA family protein |
-Macromolecule #2: Motility protein A
| Macromolecule | Name: Motility protein A / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium sporogenes (bacteria) Clostridium sporogenes (bacteria) |
| Molecular weight | Theoretical: 28.92885 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKRDILTPI GFVLCFGLVL WGMASGGSNL KVFWDVASVF ITIGGSMAAM LITYPMDEFK RLLIVIRQTF KDNGMSNIDV IQNFVDLSR KARREGLLSL EDAINNLTDD YMKKGLRMVV DGIEPETIRE IMELEIDEME KRHKSGADML KTWGGYAPAF G MVGTLIGL ...String: MKKRDILTPI GFVLCFGLVL WGMASGGSNL KVFWDVASVF ITIGGSMAAM LITYPMDEFK RLLIVIRQTF KDNGMSNIDV IQNFVDLSR KARREGLLSL EDAINNLTDD YMKKGLRMVV DGIEPETIRE IMELEIDEME KRHKSGADML KTWGGYAPAF G MVGTLIGL IQMLANLTDS STIASGMGKA LITTFYGSLM ANAVFNPMGA NLMFKSGVEA TTREMVLEGV LAIQSGVNPR IM EEKLVSY LSPPERQAYS KVQVS UniProtKB: Motility protein A |
-Macromolecule #3: Flagellar motor switch protein FliG
| Macromolecule | Name: Flagellar motor switch protein FliG / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium sporogenes (bacteria) Clostridium sporogenes (bacteria) |
| Molecular weight | Theoretical: 11.311659 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGGSGGGSGG GSVFEDIITL DDVAIQRVLR EVETKDLALA LKGSSEEVAN VIFRNQSKRA ASSLKEDIEF LGPVRIMDVE KAQQGIVSI IRRLDEAGEI VISRGGED UniProtKB: Flagellar motor switch protein FliG |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 56.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)