+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
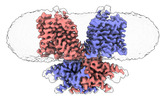
| Title | Cryo-EM structure of dolphin Prestin in low Cl buffer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Solute Carrier / Electromotility / Voltage Sensitive / Mechanosensitive / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcochlear outer hair cell electromotile response / secondary active sulfate transmembrane transporter activity / sensory perception of sound / regulation of cell shape / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
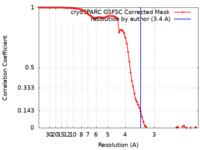
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Haller P / Bavi N / Perozo E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Folding of prestin's anion-binding site and the mechanism of outer hair cell electromotility. Authors: Xiaoxuan Lin / Patrick R Haller / Navid Bavi / Nabil Faruk / Eduardo Perozo / Tobin R Sosnick /  Abstract: Prestin responds to transmembrane voltage fluctuations by changing its cross-sectional area, a process underlying the electromotility of outer hair cells and cochlear amplification. Prestin belongs ...Prestin responds to transmembrane voltage fluctuations by changing its cross-sectional area, a process underlying the electromotility of outer hair cells and cochlear amplification. Prestin belongs to the SLC26 family of anion transporters yet is the only member capable of displaying electromotility. Prestin's voltage-dependent conformational changes are driven by the putative displacement of residue R399 and a set of sparse charged residues within the transmembrane domain, following the binding of a Cl anion at a conserved binding site formed by the amino termini of the TM3 and TM10 helices. However, a major conundrum arises as to how an anion that binds in proximity to a positive charge (R399), can promote the voltage sensitivity of prestin. Using hydrogen-deuterium exchange mass spectrometry, we find that prestin displays an unstable anion-binding site, where folding of the amino termini of TM3 and TM10 is coupled to Cl binding. This event shortens the TM3-TM10 electrostatic gap, thereby connecting the two helices, resulting in reduced cross-sectional area. These folding events upon anion binding are absent in SLC26A9, a non-electromotile transporter closely related to prestin. Dynamics of prestin embedded in a lipid bilayer closely match that in detergent micelle, except for a destabilized lipid-facing helix TM6 that is critical to prestin's mechanical expansion. We observe helix fraying at prestin's anion-binding site but cooperative unfolding of multiple lipid-facing helices, features that may promote prestin's fast electromechanical rearrangements. These results highlight a novel role of the folding equilibrium of the anion-binding site, and help define prestin's unique voltage-sensing mechanism and electromotility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42112.map.gz emd_42112.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42112-v30.xml emd-42112-v30.xml emd-42112.xml emd-42112.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42112_fsc.xml emd_42112_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42112.png emd_42112.png | 102 KB | ||
| Filedesc metadata |  emd-42112.cif.gz emd-42112.cif.gz | 6.4 KB | ||
| Others |  emd_42112_additional_1.map.gz emd_42112_additional_1.map.gz emd_42112_half_map_1.map.gz emd_42112_half_map_1.map.gz emd_42112_half_map_2.map.gz emd_42112_half_map_2.map.gz | 30.7 MB 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42112 http://ftp.pdbj.org/pub/emdb/structures/EMD-42112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42112 | HTTPS FTP |
-Validation report
| Summary document |  emd_42112_validation.pdf.gz emd_42112_validation.pdf.gz | 806.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42112_full_validation.pdf.gz emd_42112_full_validation.pdf.gz | 806.1 KB | Display | |
| Data in XML |  emd_42112_validation.xml.gz emd_42112_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_42112_validation.cif.gz emd_42112_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42112 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42112 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42112 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42112 | HTTPS FTP |
-Related structure data
| Related structure data |  8uc1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42112.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42112.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.068 Å | ||||||||||||||||||||||||||||||||||||
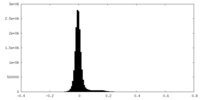
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_42112_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
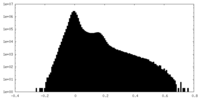
| Density Histograms |
-Half map: #2
| File | emd_42112_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_42112_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dolphin Prestin solubilized in GDN in low Cl buffer
| Entire | Name: Dolphin Prestin solubilized in GDN in low Cl buffer |
|---|---|
| Components |
|
-Supramolecule #1: Dolphin Prestin solubilized in GDN in low Cl buffer
| Supramolecule | Name: Dolphin Prestin solubilized in GDN in low Cl buffer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Prestin
| Macromolecule | Name: Prestin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.97375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDHVEETEIL AATQRYYVER PIFSHPVLQE RLHKKDKISE SIGDKLKQAF TCTPKKIRNI IYMFLPITKW LPAYRFKEYV LGDIVSGIS TGVLQLPQGL AFAMLAAVPP VFGLYSSFYP VIMYCFFGTS RHISIGPFAV ISLMIGGVAV RLVPDDIVIP G GVNATNST ...String: MDHVEETEIL AATQRYYVER PIFSHPVLQE RLHKKDKISE SIGDKLKQAF TCTPKKIRNI IYMFLPITKW LPAYRFKEYV LGDIVSGIS TGVLQLPQGL AFAMLAAVPP VFGLYSSFYP VIMYCFFGTS RHISIGPFAV ISLMIGGVAV RLVPDDIVIP G GVNATNST EARDALRVKV AMSVTLLTGI IQFCLGVCRF GFVAIYLTEP LVRGFTTAAA VHVFTSMLKY LFGVKTKRYS GI FSVVYST VAVLQNVKNL NVCSLGVGLM VFGLLLGGKE FNERFKEKLP APIPLEFFAV VMGTGISAGF SLHESYNVDV VGT LPLGLL PPANPDTSLF HLVYVDAIAI AIVGFSVTIS MAKTLANKHG YQVDGNQELI ALGLCNSTGS LFQTFAISCS LSRS LVQEG TGGKTQLAGC LASLMILLVI LATGFLFESL PQAVLSAIVI VNLKGMFMQF SDLPFFWRTS KIELTIWLTT FVSSL FLGL DYGLITAVII ALMTVIYRTQ SPSYIVLGQL PDTDVYIDID AYEEVKEVPG IKIFQINAPI YYANSDLYSS ALKRKT GVN PAFILGARRK AMKKYAKEVG NANMANATVV KVDAEVDAED GTKPEEEEDE IKYPPIVTKS TLPEELQRFM PPGDNVH TI ILDFTQVNFM DSVGVKTLAG IVKEYGDVGI YVYLAGCSAQ VVSDLTQNQF FENPALLDLL FHSIHDAVLG SQVREALA E QEATAAPPQE DSEPNATPEA UniProtKB: Prestin |
-Macromolecule #2: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 190 mM HEPES, 95 mM Tris-base, 1mM NaCl, 3mM DTT, 1mM EDTA, 0.02 % GDN | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5s Blot time, Blot force 1. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)