+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural Basis of Human NOX5 Activation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | enzyme / oxidase / activation mechanism / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of fusion of sperm to egg plasma membrane / superoxide-generating NADPH oxidase activity / Oxidoreductases; Acting on NADH or NADPH; With oxygen as acceptor / cytoskeleton-dependent cytokinesis / NADPH oxidase complex / superoxide-generating NAD(P)H oxidase activity / endothelial cell proliferation / proton channel activity / superoxide anion generation / Detoxification of Reactive Oxygen Species ...regulation of fusion of sperm to egg plasma membrane / superoxide-generating NADPH oxidase activity / Oxidoreductases; Acting on NADH or NADPH; With oxygen as acceptor / cytoskeleton-dependent cytokinesis / NADPH oxidase complex / superoxide-generating NAD(P)H oxidase activity / endothelial cell proliferation / proton channel activity / superoxide anion generation / Detoxification of Reactive Oxygen Species / proton transmembrane transport / positive regulation of cytokine production / defense response / positive regulation of reactive oxygen species metabolic process / NADP binding / flavin adenine dinucleotide binding / angiogenesis / apoptotic process / heme binding / calcium ion binding / endoplasmic reticulum membrane / endoplasmic reticulum / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.06 Å | |||||||||
 Authors Authors | Cui C / Jiang M / Sun J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of human NOX5 activation. Authors: Chenxi Cui / Meiqin Jiang / Nikhil Jain / Sourav Das / Yu-Hua Lo / Ali A Kermani / Tanadet Pipatpolkai / Ji Sun /   Abstract: NADPH oxidase 5 (NOX5) catalyzes the production of superoxide free radicals and regulates physiological processes from sperm motility to cardiac rhythm. Overexpression of NOX5 leads to cancers, ...NADPH oxidase 5 (NOX5) catalyzes the production of superoxide free radicals and regulates physiological processes from sperm motility to cardiac rhythm. Overexpression of NOX5 leads to cancers, diabetes, and cardiovascular diseases. NOX5 is activated by intracellular calcium signaling, but the underlying molecular mechanism of which - in particular, how calcium triggers electron transfer from NADPH to FAD - is still unclear. Here we capture motions of full-length human NOX5 upon calcium binding using single-particle cryogenic electron microscopy (cryo-EM). By combining biochemistry, mutagenesis analyses, and molecular dynamics (MD) simulations, we decode the molecular basis of NOX5 activation and electron transfer. We find that calcium binding to the EF-hand domain increases NADPH dynamics, permitting electron transfer between NADPH and FAD and superoxide production. Our structural findings also uncover a zinc-binding motif that is important for NOX5 stability and enzymatic activity, revealing modulation mechanisms of reactive oxygen species (ROS) production. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42013.map.gz emd_42013.map.gz | 43.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42013-v30.xml emd-42013-v30.xml emd-42013.xml emd-42013.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42013.png emd_42013.png | 75.2 KB | ||
| Filedesc metadata |  emd-42013.cif.gz emd-42013.cif.gz | 6.2 KB | ||
| Others |  emd_42013_additional_1.map.gz emd_42013_additional_1.map.gz emd_42013_additional_2.map.gz emd_42013_additional_2.map.gz emd_42013_additional_3.map.gz emd_42013_additional_3.map.gz emd_42013_additional_4.map.gz emd_42013_additional_4.map.gz emd_42013_half_map_1.map.gz emd_42013_half_map_1.map.gz emd_42013_half_map_2.map.gz emd_42013_half_map_2.map.gz | 40.4 MB 39.8 MB 39.8 MB 49.7 MB 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42013 http://ftp.pdbj.org/pub/emdb/structures/EMD-42013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42013 | HTTPS FTP |
-Related structure data
| Related structure data |  8u7yMC  8u85C  8u86C  8u87C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42013.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42013.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.297 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #2
| File | emd_42013_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
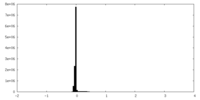
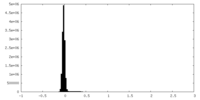
| Density Histograms |
-Additional map: #3
| File | emd_42013_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
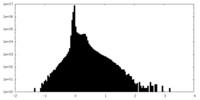
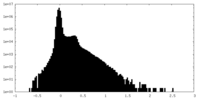
| Density Histograms |
-Additional map: #4
| File | emd_42013_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
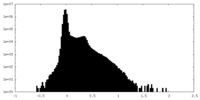
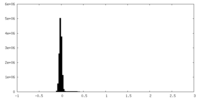
| Density Histograms |
-Additional map: #1
| File | emd_42013_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
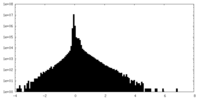
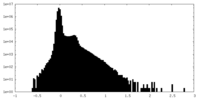
| Density Histograms |
-Half map: #1
| File | emd_42013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NADPH oxidase 5
| Entire | Name: NADPH oxidase 5 |
|---|---|
| Components |
|
-Supramolecule #1: NADPH oxidase 5
| Supramolecule | Name: NADPH oxidase 5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NADPH oxidase 5
| Macromolecule | Name: NADPH oxidase 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Oxidoreductases; Acting on NADH or NADPH; With oxygen as acceptor |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.118992 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAEEDARWL RWVTQQFKTI AGEDGEISLQ EFKAALHVKE SFFAERFFAL FDSDRSGTIT LQELQEALTL LIHGSPMDKL KFLFQVYDI DGSGSIDPDE LRTVLQSCLR ESAISLPDEK LDQLTLALFE SADADGNGAI TFEELRDELQ RFPGVMENLT I SAAHWLTA ...String: MSAEEDARWL RWVTQQFKTI AGEDGEISLQ EFKAALHVKE SFFAERFFAL FDSDRSGTIT LQELQEALTL LIHGSPMDKL KFLFQVYDI DGSGSIDPDE LRTVLQSCLR ESAISLPDEK LDQLTLALFE SADADGNGAI TFEELRDELQ RFPGVMENLT I SAAHWLTA PAPRPRPRRP RQLTRAYWHN HRSQLFCLAT YAGLHVLLFG LAASAHRDLG ASVMVAKGCG QCLNFDCSFI AV LMLRRCL TWLRATWLAQ VLPLDQNIQF HQLMGYVVVG LSLVHTVAHT VNFVLQAQAE ASPFQFWELL LTTRPGIGWV HGS ASPTGV ALLLLLLLMF ICSSSCIRRS GHFEVFYWTH LSYLLVWLLL IFHGPNFWKW LLVPGILFFL EKAIGLAVSR MAAV CIMEV NLLPSKVTHL LIKRPPFFHY RPGDYLYLNI PTIARYEWHP FTISSAPEQK DTIWLHIRSQ GQWTNRLYES FKASD PLGR GSKRLSRSVT MRKSQRSSKG SEILLEKHKF CNIKCYIDGP YGTPTRRIFA SEHAVLIGAG IGITPFASIL QSIMYR HQK RKHTCPSCQH SWIEGVQDNM KLHKVDFIWI NRDQRSFEWF VSLLTKLEMD QAEEAQYGRF LELHMYMTSA LGKNDMK AI GLQMALDLLA NKEKKDSITG LQTRTQPGRP DWSKVFQKVA AEKKGKVQVF FCGSPALAKV LKGHCEKFGF RFFQENF UniProtKB: NADPH oxidase 5 |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 3 / Number of copies: 2 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #4: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 4 / Number of copies: 2 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #5: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAP |
|---|---|
| Molecular weight | Theoretical: 743.405 Da |
| Chemical component information |  ChemComp-NAP: |
-Macromolecule #6: HEME B/C
| Macromolecule | Name: HEME B/C / type: ligand / ID: 6 / Number of copies: 4 / Formula: HEB |
|---|---|
| Molecular weight | Theoretical: 618.503 Da |
| Chemical component information |  ChemComp-HEB: |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.06 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 220000 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)