[English] 日本語
 Yorodumi
Yorodumi- EMDB-41983: Structure of the phage immune evasion protein Gad1 bound to the G... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | ||||||||||||||||||
 Map data Map data | Main cryo-EM Map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | viral immune evasion / phage / bacteria / anti-phage defense complex / DNA nuclease / DNA helicase / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationrecombinational repair / 3'-5' DNA helicase activity / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / symbiont-mediated suppression of host innate immune response / hydrolase activity / DNA binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Bacillus phage phi3T (virus) / Bacillus phage phi3T (virus) /  | ||||||||||||||||||
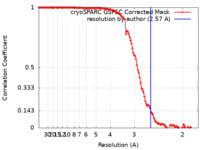
| Method | single particle reconstruction / cryo EM / Resolution: 2.57 Å | ||||||||||||||||||
 Authors Authors | Antine SP / Johnson AG / Mooney SE / Mayer ML / Kranzsuch PJ | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural basis of Gabija anti-phage defence and viral immune evasion. Authors: Sadie P Antine / Alex G Johnson / Sarah E Mooney / Azita Leavitt / Megan L Mayer / Erez Yirmiya / Gil Amitai / Rotem Sorek / Philip J Kranzusch /   Abstract: Bacteria encode hundreds of diverse defence systems that protect them from viral infection and inhibit phage propagation. Gabija is one of the most prevalent anti-phage defence systems, occurring in ...Bacteria encode hundreds of diverse defence systems that protect them from viral infection and inhibit phage propagation. Gabija is one of the most prevalent anti-phage defence systems, occurring in more than 15% of all sequenced bacterial and archaeal genomes, but the molecular basis of how Gabija defends cells from viral infection remains poorly understood. Here we use X-ray crystallography and cryo-electron microscopy (cryo-EM) to define how Gabija proteins assemble into a supramolecular complex of around 500 kDa that degrades phage DNA. Gabija protein A (GajA) is a DNA endonuclease that tetramerizes to form the core of the anti-phage defence complex. Two sets of Gabija protein B (GajB) dimers dock at opposite sides of the complex and create a 4:4 GajA-GajB assembly (hereafter, GajAB) that is essential for phage resistance in vivo. We show that a phage-encoded protein, Gabija anti-defence 1 (Gad1), directly binds to the Gabija GajAB complex and inactivates defence. A cryo-EM structure of the virally inhibited state shows that Gad1 forms an octameric web that encases the GajAB complex and inhibits DNA recognition and cleavage. Our results reveal the structural basis of assembly of the Gabija anti-phage defence complex and define a unique mechanism of viral immune evasion. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Phages overcome bacterial immunity via diverse anti-defense proteins Authors: Yirmiya E / Leavitt A / Lu A / Avraham C / Osterman I / Garb J / Antine SP / Mooney SE / Hobbs SJ / Kranzusch PJ / Amitai G / Sorek R | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41983.map.gz emd_41983.map.gz | 80.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41983-v30.xml emd-41983-v30.xml emd-41983.xml emd-41983.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41983_fsc.xml emd_41983_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41983.png emd_41983.png | 96.6 KB | ||
| Filedesc metadata |  emd-41983.cif.gz emd-41983.cif.gz | 6.8 KB | ||
| Others |  emd_41983_half_map_1.map.gz emd_41983_half_map_1.map.gz emd_41983_half_map_2.map.gz emd_41983_half_map_2.map.gz | 151.7 MB 151.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41983 http://ftp.pdbj.org/pub/emdb/structures/EMD-41983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41983 | HTTPS FTP |
-Related structure data
| Related structure data |  8u7iMC  8sm3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41983.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41983.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main cryo-EM Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B
| File | emd_41983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_41983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phage Phi3T Gad1 bound to the Bacillus cereus Gabija GajAB complex
| Entire | Name: Phage Phi3T Gad1 bound to the Bacillus cereus Gabija GajAB complex |
|---|---|
| Components |
|
-Supramolecule #1: Phage Phi3T Gad1 bound to the Bacillus cereus Gabija GajAB complex
| Supramolecule | Name: Phage Phi3T Gad1 bound to the Bacillus cereus Gabija GajAB complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bacillus phage phi3T (virus) Bacillus phage phi3T (virus) |
| Molecular weight | Theoretical: 280 KDa |
-Supramolecule #2: Gabija GajAB complex
| Supramolecule | Name: Gabija GajAB complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Gad1 Octamer
| Supramolecule | Name: Gad1 Octamer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Bacillus phage phi3T (virus) Bacillus phage phi3T (virus) |
-Macromolecule #1: Endonuclease GajA
| Macromolecule | Name: Endonuclease GajA / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.045656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH GSGVKTENND HINLKVAGQD GSVVQFKIKR HTPLSKLMKA YCERQGLSMR QIRFRFDGQP INETDTPAQL EMEDEDTID VFQQQTGGSK FSNITIKNFR NFEKVNINLD NKNVIFGMND IGKTNFLYAL RFLLDKEIRK FGFNKSDYHK H DTSKKIEI ...String: MGSSHHHHHH GSGVKTENND HINLKVAGQD GSVVQFKIKR HTPLSKLMKA YCERQGLSMR QIRFRFDGQP INETDTPAQL EMEDEDTID VFQQQTGGSK FSNITIKNFR NFEKVNINLD NKNVIFGMND IGKTNFLYAL RFLLDKEIRK FGFNKSDYHK H DTSKKIEI ILTLDLSNYE KDEDTKKLIS VVKGARTSAN ADVFYIALES KYDDKELYGN IILKWGSELD NLIDIPGRGN IN ALDNVFK VIYINPLVDL DKLFAQNKKY IFEESQGNES DEGILNNIKS LTDQVNQQIG EMTIIKGFQQ EITSEYRSLK KEE VSIELK SEMAIKGFFS DIIPYIKKDG DSNYYPTSGD GRRKMLSYSI YNYLAKKKYE DKIVIYLIEE PEISLHRSMQ IALS KQLFE QSTYKYFFLS THSPELLYEM DNTRLIRVHS TEKVVCSSHM YNVEEAYGSV KKKLNKALSS ALFAERVLLI EGPSE KILF EKVLDEVEPE YELNGGFLLE VGGTYFNHYV CTLNDLGITH IIKTDNDLKS KKGKKGVYEL LGLNRCLNLL GRENLD EIT IDIPEDIKGK KKKERLNERK KEIFKQYKNE VGEFLGERIY LSEIDLENDL YSAIGESMKR IFENEDPVHY LQKSKLF NM VELVNNLSTK DCFDVFEHEK FACLKELVGS DRG UniProtKB: Endonuclease GajA |
-Macromolecule #2: Gabija protein GajB
| Macromolecule | Name: Gabija protein GajB / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.139992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSREQIIKDG GNILVTAGAG SGKTTILVSK IEADLKENKT HYSIAAVTFT NKAAKEIEGR LGYSSRGNFI GTNDGFVESE IIRPFIKDA FGNDYPDNFT AEYFDNQFAS YDKGLQVLKY QNILGTYSNP KKNFKFQLAL DILKKSLVAR QYIFSKYFKI F IDEYQDSD ...String: MSREQIIKDG GNILVTAGAG SGKTTILVSK IEADLKENKT HYSIAAVTFT NKAAKEIEGR LGYSSRGNFI GTNDGFVESE IIRPFIKDA FGNDYPDNFT AEYFDNQFAS YDKGLQVLKY QNILGTYSNP KKNFKFQLAL DILKKSLVAR QYIFSKYFKI F IDEYQDSD KDMHNLFMYL KDQLKIKLFI VGDPKQSIYI WRGAEPENFN GLIENSTDFN KYHLTSNFRC CQDIQNYSNL FN EETRSLI KEKNEVQNVI SIADDMPISD ILLKLTEEKQ VLNIEAELVI LVRRRNQAIE IMKELNEEGF NFIFIPQTPL DRA TPNATL LKEVIKYVKN DRYSIYDLAA EIVGNLSSRE IKEIQKIINE LLVPNINQVL INQVLINLFA KLEITLDTRE ITAF TEVMM TNEFDIAFDT NEYLHKIFTV HSAKGLEFNQ VIITASDYNV HYNRDTNEHY VATTRAKDKL IVIMDNKKYS DYIET LMKE LKIKNIIKSI UniProtKB: Gabija protein GajB |
-Macromolecule #3: Gabija Anti-Defense 1
| Macromolecule | Name: Gabija Anti-Defense 1 / type: protein_or_peptide / ID: 3 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillus phage phi3T (virus) Bacillus phage phi3T (virus) |
| Molecular weight | Theoretical: 34.919184 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLIGIKTSN CFLVSDNIEG KRYFHSQLDE LLFDGKRATE TYKSDWFKLE KEPSVIEKQM PAKKINHRYE LKEGFQESEL TPKVIKASY IGEDSEYYEV KGLYDLKFEE IPQQNEKIEF EMNVIEEIDG ELKLQSHNFN LNYNLLDRIQ THPMLLETKP C YLSQEESY ...String: MKLIGIKTSN CFLVSDNIEG KRYFHSQLDE LLFDGKRATE TYKSDWFKLE KEPSVIEKQM PAKKINHRYE LKEGFQESEL TPKVIKASY IGEDSEYYEV KGLYDLKFEE IPQQNEKIEF EMNVIEEIDG ELKLQSHNFN LNYNLLDRIQ THPMLLETKP C YLSQEESY KIIRNHIKAN INPKFARITS DYDFCLTVVK VLELYKPHEY IVDLNAMYKR RKPKLEKRFQ TKREVEIYKV AP KAYQSYP IVEPFSGKDV EDLKSNIKKF LDDLMAKINE PLVECKCCKG RGVILNEN UniProtKB: Gabija anti-defense 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 1.00 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES-KOH pH 7.5, 20 mM KCl, and 1 mM TCEP-KOH |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 41.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)