[English] 日本語
 Yorodumi
Yorodumi- EMDB-41919: Structure of E.coli beta-Galactosidase from a multi-species dataset -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E.coli beta-Galactosidase from a multi-species dataset | |||||||||
 Map data Map data | Final map sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hydrolase / D2 / beta-gal | |||||||||
| Biological species |  | |||||||||
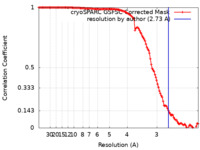
| Method | single particle reconstruction / cryo EM / Resolution: 2.73 Å | |||||||||
 Authors Authors | Kopylov M / Bobe D / Eng ET | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr F Struct Biol Commun / Year: 2024 Journal: Acta Crystallogr F Struct Biol Commun / Year: 2024Title: Multi-species cryoEM calibration and workflow verification standard. Authors: Daija Bobe / Mykhailo Kopylov / Jessalyn Miller / Aaron P Owji / Edward T Eng /  Abstract: Cryogenic electron microscopy (cryoEM) is a rapidly growing structural biology modality that has been successful in revealing molecular details of biological systems. However, unlike established ...Cryogenic electron microscopy (cryoEM) is a rapidly growing structural biology modality that has been successful in revealing molecular details of biological systems. However, unlike established biophysical and analytical techniques with calibration standards, cryoEM has lacked comprehensive biological test samples. Here, a cryoEM calibration sample consisting of a mixture of compatible macromolecules is introduced that can not only be used for resolution optimization, but also provides multiple reference points for evaluating instrument performance, data quality and image-processing workflows in a single experiment. This combined test specimen provides researchers with a reference point for validating their cryoEM pipeline, benchmarking their methodologies and testing new algorithms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41919.map.gz emd_41919.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41919-v30.xml emd-41919-v30.xml emd-41919.xml emd-41919.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41919_fsc.xml emd_41919_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_41919.png emd_41919.png | 143.4 KB | ||
| Masks |  emd_41919_msk_1.map emd_41919_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41919.cif.gz emd-41919.cif.gz | 4.3 KB | ||
| Others |  emd_41919_half_map_1.map.gz emd_41919_half_map_1.map.gz emd_41919_half_map_2.map.gz emd_41919_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41919 http://ftp.pdbj.org/pub/emdb/structures/EMD-41919 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41919 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41919 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41919.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41919.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map sharp | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41919_msk_1.map emd_41919_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
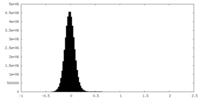
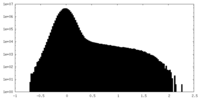
| Density Histograms |
-Half map: Halfmap A
| File | emd_41919_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap B
| File | emd_41919_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : beta-Galactosidase from Escherichia coli in complex with 2-phenyl...
| Entire | Name: beta-Galactosidase from Escherichia coli in complex with 2-phenylethyl beta-D-thiogalactoside |
|---|---|
| Components |
|
-Supramolecule #1: beta-Galactosidase from Escherichia coli in complex with 2-phenyl...
| Supramolecule | Name: beta-Galactosidase from Escherichia coli in complex with 2-phenylethyl beta-D-thiogalactoside type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)