+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Klebsiella pneumoniae encapsulin-associated DyP peroxidase | |||||||||
 Map data Map data | Klebsiella pneumoniae encapsulin-associated DyP peroxidase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DyP peroxidase / encapsulin / OXIDOREDUCTASE | |||||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.39 Å | |||||||||
 Authors Authors | Andreas MP / Jones JA / Giessen TW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for peroxidase encapsulation inside the encapsulin from the Gram-negative pathogen Klebsiella pneumoniae. Authors: Jesse A Jones / Michael P Andreas / Tobias W Giessen /  Abstract: Encapsulins are self-assembling protein nanocompartments capable of selectively encapsulating dedicated cargo proteins, including enzymes involved in iron storage, sulfur metabolism, and stress ...Encapsulins are self-assembling protein nanocompartments capable of selectively encapsulating dedicated cargo proteins, including enzymes involved in iron storage, sulfur metabolism, and stress resistance. They represent a unique compartmentalization strategy used by many pathogens to facilitate specialized metabolic capabilities. Encapsulation is mediated by specific cargo protein motifs known as targeting peptides (TPs), though the structural basis for encapsulation of the largest encapsulin cargo class, dye-decolorizing peroxidases (DyPs), is currently unknown. Here, we characterize a DyP-containing encapsulin from the enterobacterial pathogen Klebsiella pneumoniae. By combining cryo-electron microscopy with TP and TP-binding site mutagenesis, we elucidate the molecular basis for cargo encapsulation. TP binding is mediated by cooperative hydrophobic and ionic interactions as well as shape complementarity. Our results expand the molecular understanding of enzyme encapsulation inside protein nanocompartments and lay the foundation for rationally modulating encapsulin cargo loading for biomedical and biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41904.map.gz emd_41904.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41904-v30.xml emd-41904-v30.xml emd-41904.xml emd-41904.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
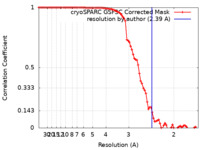
| FSC (resolution estimation) |  emd_41904_fsc.xml emd_41904_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_41904.png emd_41904.png | 129.9 KB | ||
| Filedesc metadata |  emd-41904.cif.gz emd-41904.cif.gz | 6.5 KB | ||
| Others |  emd_41904_half_map_1.map.gz emd_41904_half_map_1.map.gz emd_41904_half_map_2.map.gz emd_41904_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41904 http://ftp.pdbj.org/pub/emdb/structures/EMD-41904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41904 | HTTPS FTP |
-Related structure data
| Related structure data |  8u4zMC  8u50C  8u51C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41904.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41904.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Klebsiella pneumoniae encapsulin-associated DyP peroxidase | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map B
| File | emd_41904_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
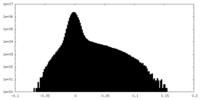
| Projections & Slices |
| ||||||||||||
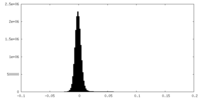
| Density Histograms |
-Half map: Half Map A
| File | emd_41904_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
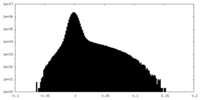
| Projections & Slices |
| ||||||||||||
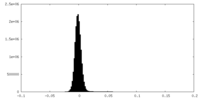
| Density Histograms |
- Sample components
Sample components
-Entire : Klebsiella pneumoniae family 1 encapsulin-associated DyP peroxidase
| Entire | Name: Klebsiella pneumoniae family 1 encapsulin-associated DyP peroxidase |
|---|---|
| Components |
|
-Supramolecule #1: Klebsiella pneumoniae family 1 encapsulin-associated DyP peroxidase
| Supramolecule | Name: Klebsiella pneumoniae family 1 encapsulin-associated DyP peroxidase type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Family 1 encapsulin-associated DyP peroxidase
| Macromolecule | Name: Family 1 encapsulin-associated DyP peroxidase / type: protein_or_peptide / ID: 1 Details: Residues 1-352 are K.p. DyP peroxidase (NCBI: WP_193221875.1). Residues 1-2 and 316-352 are disordered. Residues 353-370 are a TEV protease site, linker, and HIS-tag that are disordered in the structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 40.691395 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MACPISQSVS QPVDERLTRA AIFLVVTINP GKAAEVAVRT LCGTLSSLVR GVGFRILDGG LSCVMGVSSG GWERLFGDEK PEYLHVFQE INGVHHAPST PGDLLFHIRA ARMDLCFELA SRILSDLGSS VCVVDSVQGF RYFDDRDLLG FVDGTENPVA Q AAVDATLI ...String: MACPISQSVS QPVDERLTRA AIFLVVTINP GKAAEVAVRT LCGTLSSLVR GVGFRILDGG LSCVMGVSSG GWERLFGDEK PEYLHVFQE INGVHHAPST PGDLLFHIRA ARMDLCFELA SRILSDLGSS VCVVDSVQGF RYFDDRDLLG FVDGTENPVA Q AAVDATLI GEEDHTFSGG SYVIVQKYLH DLDKWNAIPV EQQEKIIGRE KLSDIELKDA DKPSYAHNVL TSIEEDGEDV DI LRDNMPF GDPGKGEFGT YFIGYSRKPA RIERMLENMF VGNPPGNYDR ILDVSRAITG TLFFIPTVSF LDSVEPQPSV SQQ TDDAKY IYDSPGTKGE SGSLNIGSLK KEVQDEENLY FQGGSGGHHH HHHH |
-Macromolecule #2: Mesoheme
| Macromolecule | Name: Mesoheme / type: ligand / ID: 2 / Number of copies: 1 / Formula: MH0 |
|---|---|
| Molecular weight | Theoretical: 620.519 Da |
| Chemical component information |  ChemComp-MH0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM Tris pH 7.5, 150 mM NaCl | |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 60 seconds at 5 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force: 5 Blot time: 2 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2468 / Average exposure time: 1.89747 sec. / Average electron dose: 50.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: Other / Chain - Initial model type: in silico model Details: An AlphaFill model containing bound heme was used as starting model |
|---|---|
| Details | Initial fitting was performed using ChimeraX v1.2.5. The model was then manually refined and mutated using Coot v9.8.1 followed by real-space refinement using Phenix v1.20.1-4487-000. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 107.7 / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8u4z: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)