[English] 日本語
 Yorodumi
Yorodumi- EMDB-41839: Cryo-EM structure of yeast SWR1C subunit Swc5 bound to the nucleo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast SWR1C subunit Swc5 bound to the nucleosome, 3D class 0 | |||||||||||||||
 Map data Map data | Map of 3D class 0 (Fig. 6B, S1E) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Chromatin / Remodeler / Complex / Histone / GENE REGULATION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHATs acetylate histones / Condensation of Prophase Chromosomes / : / : / Assembly of the ORC complex at the origin of replication / HDACs deacetylate histones / Swr1 complex / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / Oxidative Stress Induced Senescence / RMTs methylate histone arginines ...HATs acetylate histones / Condensation of Prophase Chromosomes / : / : / Assembly of the ORC complex at the origin of replication / HDACs deacetylate histones / Swr1 complex / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / Oxidative Stress Induced Senescence / RMTs methylate histone arginines / DNA damage tolerance / RNA Polymerase I Promoter Escape / Estrogen-dependent gene expression / Ub-specific processing proteases / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / chromatin organization / chromatin remodeling / protein heterodimerization activity / DNA repair / regulation of DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
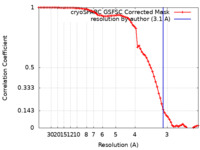
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Eek P / Tan S | |||||||||||||||
| Funding support |  United States, United States,  Estonia, 4 items Estonia, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Dual engagement of the nucleosomal acidic patches is essential for deposition of histone H2A.Z by SWR1C. Authors: Alexander S Baier / Nathan Gioacchini / Priit Eek / Erik M Leith / Song Tan / Craig L Peterson /   Abstract: The yeast SWR1C chromatin remodeling enzyme catalyzes the ATP-dependent exchange of nucleosomal histone H2A for the histone variant H2A.Z, a key variant involved in a multitude of nuclear functions. ...The yeast SWR1C chromatin remodeling enzyme catalyzes the ATP-dependent exchange of nucleosomal histone H2A for the histone variant H2A.Z, a key variant involved in a multitude of nuclear functions. How the 14-subunit SWR1C engages the nucleosomal substrate remains largely unknown. Studies on the ISWI, CHD1, and SWI/SNF families of chromatin remodeling enzymes have demonstrated key roles for the nucleosomal acidic patch for remodeling activity, however a role for this nucleosomal epitope in nucleosome editing by SWR1C has not been tested. Here, we employ a variety of biochemical assays to demonstrate an essential role for the acidic patch in the H2A.Z exchange reaction. Utilizing asymmetrically assembled nucleosomes, we demonstrate that the acidic patches on each face of the nucleosome are required for SWR1C-mediated dimer exchange, suggesting SWR1C engages the nucleosome in a 'pincer-like' conformation, engaging both patches simultaneously. Loss of a single acidic patch results in loss of high affinity nucleosome binding and nucleosomal stimulation of ATPase activity. We identify a conserved arginine-rich motif within the Swc5 subunit that binds the acidic patch and is key for dimer exchange activity. In addition, our cryoEM structure of a Swc5-nucleosome complex suggests that promoter proximal, histone H2B ubiquitylation may regulate H2A.Z deposition. Together these findings provide new insights into how SWR1C engages its nucleosomal substrate to promote efficient H2A.Z deposition. #1:  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Dual engagement of the nucleosomal acidic patches is essential for deposition of histone H2A.Z by SWR1C Authors: Baier AS / Gioacchini N / Eek P / Leith EM / Song T / Peterson CL | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41839.map.gz emd_41839.map.gz | 19.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41839-v30.xml emd-41839-v30.xml emd-41839.xml emd-41839.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41839_fsc.xml emd_41839_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_41839.png emd_41839.png | 180.4 KB | ||
| Masks |  emd_41839_msk_1.map emd_41839_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41839.cif.gz emd-41839.cif.gz | 6.3 KB | ||
| Others |  emd_41839_half_map_1.map.gz emd_41839_half_map_1.map.gz emd_41839_half_map_2.map.gz emd_41839_half_map_2.map.gz | 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41839 http://ftp.pdbj.org/pub/emdb/structures/EMD-41839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41839 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41839.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41839.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of 3D class 0 (Fig. 6B, S1E) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.182 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41839_msk_1.map emd_41839_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of class 0
| File | emd_41839_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of class 0 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of class 0
| File | emd_41839_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of class 0 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Swc5 in complex with nucleosome core particle
| Entire | Name: Swc5 in complex with nucleosome core particle |
|---|---|
| Components |
|
-Supramolecule #1: Swc5 in complex with nucleosome core particle
| Supramolecule | Name: Swc5 in complex with nucleosome core particle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Histone H3.2
| Macromolecule | Name: Histone H3.2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAVM ALQEASEAYL VALFEDTNLC AIHAKRVTIM PKDIQLARRI RGERA UniProtKB: Histone H3.2 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A.1
| Macromolecule | Name: Histone H2A.1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: SGGKGGKAGS AAKASQSRSA KAGLTFPVGR VHRLLRRGNY AQRIGSGAPV YLTAVLEYLA AEILELAGNA ARDNKKTRII PRHLQLAIRN DDELNKLLGN VTIAQGGVLP NIHQNLLPKK SAKATKASQE L UniProtKB: Histone H2A.1 |
-Macromolecule #4: Histone H2B.1
| Macromolecule | Name: Histone H2B.1 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: SAKAEKKPAS KAPAEKKPAA KKTSTSTDGK KRSKARKETY SSYIYKVLKQ THPDTGISQK SMSILNSFVN DIFERIATEA SKLAAYNKKS TISAREIQTA VRLILPGELA KHAVSEGTRA VTKYSSSTQA UniProtKB: Histone H2B.1 |
-Macromolecule #5: SWR1-complex protein 5
| Macromolecule | Name: SWR1-complex protein 5 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: RGSHHHHHHG SKTRRARQAE EEYAKTHKYE SLTVESIPAK VNSIWEELQE ASKNRLLSSS GKVGSVLDGS KEARSTTAAQ QEDKILIERN YKFAGETVHE KKWVSRSSAE GQEYLNSLKF KQQAPAAPVQ LEKAVRTKSN ESRQHLRRPL KRPPLLEQII SGGLRPKLTT ...String: RGSHHHHHHG SKTRRARQAE EEYAKTHKYE SLTVESIPAK VNSIWEELQE ASKNRLLSSS GKVGSVLDGS KEARSTTAAQ QEDKILIERN YKFAGETVHE KKWVSRSSAE GQEYLNSLKF KQQAPAAPVQ LEKAVRTKSN ESRQHLRRPL KRPPLLEQII SGGLRPKLTT LEKSQLDWAS YVDRAGLNDE LVLHNKDGFL ARQEFLQRVG SAEDERYKEL RRQQLAQQLQ QDNEAS UniProtKB: SWR1-complex protein 5 |
-Macromolecule #6: Widom 601 forward DNA
| Macromolecule | Name: Widom 601 forward DNA / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: ATCGAGAATC CCGGTGCCGA GGCCGCTCAA TTGGTCGTAG ACAGCTCTAG CACCGCTTAA ACGCACGTAC GCGCTGTCCC CCGCGTTTTA ACCGCCAAGG GGATTACTCC CTAGTCTCCA GGCACGTGTC AGATATATAC ATCCGAT |
-Macromolecule #7: Widom 601 reverse DNA
| Macromolecule | Name: Widom 601 reverse DNA / type: dna / ID: 7 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: ATCGGATGTA TATATCTGAC ACGTGCCTGG AGACTAGGGA GTAATCCCCT TGGCGGTTAA AACGCGGGGG ACAGCGCGTA CGTGCGTTTA AGCGGTGCTA GAGCTGTCTA CGACCAATTG AGCGGCCTCG GCACCGGGAT TCTCGAT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA with PELCO easiGlow | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Chemically cross-linked with glutaraldehyde using the GraFix method. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 13855 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 29000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)