+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Open, inward-facing MsbA structure (OIF1) | ||||||||||||||||||
 Map data Map data | Sharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | ABC transporter / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / ATPase-coupled lipid transmembrane transporter activity / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
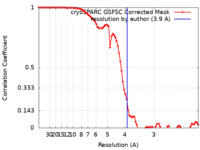
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||||||||
 Authors Authors | Yang B / Zhang T / Lyu J / Laganowsky AD / Zhao M | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Native mass spectrometry captures snapshots of the MsbA transport cycle Authors: Zhang T / Lyu J / Yang B / Zhao M / Laganowsky AD | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41597.map.gz emd_41597.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41597-v30.xml emd-41597-v30.xml emd-41597.xml emd-41597.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41597_fsc.xml emd_41597_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_41597.png emd_41597.png | 84.6 KB | ||
| Masks |  emd_41597_msk_1.map emd_41597_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41597.cif.gz emd-41597.cif.gz | 5.6 KB | ||
| Others |  emd_41597_additional_1.map.gz emd_41597_additional_1.map.gz emd_41597_half_map_1.map.gz emd_41597_half_map_1.map.gz emd_41597_half_map_2.map.gz emd_41597_half_map_2.map.gz | 31.7 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41597 http://ftp.pdbj.org/pub/emdb/structures/EMD-41597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41597 | HTTPS FTP |
-Validation report
| Summary document |  emd_41597_validation.pdf.gz emd_41597_validation.pdf.gz | 829.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41597_full_validation.pdf.gz emd_41597_full_validation.pdf.gz | 829.2 KB | Display | |
| Data in XML |  emd_41597_validation.xml.gz emd_41597_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_41597_validation.cif.gz emd_41597_validation.cif.gz | 20.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41597 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41597 | HTTPS FTP |
-Related structure data
| Related structure data |  8tspMC  8tsoC  8tsqC  8tsrC  8tssC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41597.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41597.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41597_msk_1.map emd_41597_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
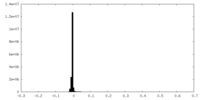
| Density Histograms |
-Additional map: unsharpened map
| File | emd_41597_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
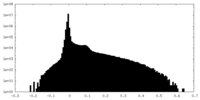
| Density Histograms |
-Half map: #2
| File | emd_41597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
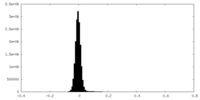
| Density Histograms |
-Half map: #1
| File | emd_41597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
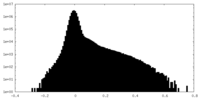
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli MsbA incubated with ATP for 6 hours
| Entire | Name: E. coli MsbA incubated with ATP for 6 hours |
|---|---|
| Components |
|
-Supramolecule #1: E. coli MsbA incubated with ATP for 6 hours
| Supramolecule | Name: E. coli MsbA incubated with ATP for 6 hours / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-binding transport protein MsbA
| Macromolecule | Name: ATP-binding transport protein MsbA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.543473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHNDKDLST WQTFRRLWPT IAPFKAGLIV AGVALILNAA SDTFMLSLLK PLLDDGFGKT DRSVLVWMPL VVIGLMILRG ITSYVSSYC ISWVSGKVVM TMRRRLFGHM MGMPVSFFDK QSTGTLLSRI TYDSEQVASS SSGALITVVR EGASIIGLFI M MFYYSWQL ...String: GSHNDKDLST WQTFRRLWPT IAPFKAGLIV AGVALILNAA SDTFMLSLLK PLLDDGFGKT DRSVLVWMPL VVIGLMILRG ITSYVSSYC ISWVSGKVVM TMRRRLFGHM MGMPVSFFDK QSTGTLLSRI TYDSEQVASS SSGALITVVR EGASIIGLFI M MFYYSWQL SIILIVLAPI VSIAIRVVSK RFRNISKNMQ NTMGQVTTSA EQMLKGHKEV LIFGGQEVET KRFDKVSNRM RL QGMKMVS ASSISDPIIQ LIASLALAFV LYAASFPSVM DSLTAGTITV VFSSMIALMR PLKSLTNVNA QFQRGMAACQ TLF TILDSE QEKDEGKRVI ERATGDVEFR NVTFTYPGRD VPALRNINLK IPAGKTVALV GRSGSGKSTI ASLITRFYDI DEGE ILMDG HDLREYTLAS LRNQVALVSQ NVHLFNDTVA NNIAYARTEQ YSREQIEEAA RMAYAMDFIN KMDNGLDTVI GENGV LLSG GQRQRIAIAR ALLRDSPILI LDEATSALDT ESERAIQAAL DELQKNRTSL VIAHRLSTIE KADEIVVVED GVIVER GTH NDLLEHRGVY AQLHKMQFGQ UniProtKB: ATP-binding transport protein MsbA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8tsp: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)