+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Fab HC84.26-HCV E2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HCV / E2 / Fab / antiviral / complex / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...host cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / virion membrane / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Hepacivirus C Hepacivirus C | |||||||||
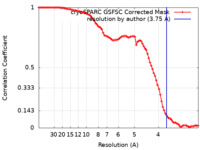
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Shahid S / Liqun J / LIu Y / Hasan SS / Mariuzza RA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2025 Journal: Commun Biol / Year: 2025Title: Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers. Authors: Salman Shahid / Sharanbasappa S Karade / S Saif Hasan / Rui Yin / Liqun Jiang / Yanxin Liu / Nathaniel Felbinger / Liudmila Kulakova / Eric A Toth / Zhen-Yong Keck / Steven K H Foung / ...Authors: Salman Shahid / Sharanbasappa S Karade / S Saif Hasan / Rui Yin / Liqun Jiang / Yanxin Liu / Nathaniel Felbinger / Liudmila Kulakova / Eric A Toth / Zhen-Yong Keck / Steven K H Foung / Thomas R Fuerst / Brian G Pierce / Roy A Mariuzza /  Abstract: Global elimination of hepatitis C virus (HCV) will require an effective cross-genotype vaccine. The HCV E2 envelope glycoprotein is the main target of neutralizing antibodies but also contains ...Global elimination of hepatitis C virus (HCV) will require an effective cross-genotype vaccine. The HCV E2 envelope glycoprotein is the main target of neutralizing antibodies but also contains epitopes that elicit non-neutralizing antibodies which may provide protection through Fc effector functions rather than direct neutralization. We determined cryo-EM structures of a broadly neutralizing antibody, a moderately neutralizing antibody, and a non-neutralizing antibody bound to E2 to resolutions of 3.8, 3.3, and 3.7 Å, respectively. Whereas the broadly neutralizing antibody targeted the front layer of E2 and the non-neutralizing antibody targeted the back layer, the moderately neutralizing antibody straddled both front and back layers, and thereby defined a new neutralizing epitope on E2. The small size of complexes between conventional (monovalent) Fabs and E2 (~110 kDa) presented a challenge for cryo-EM. Accordingly, we engineered bivalent versions of E2-specific Fabs that doubled the size of Fab-E2 complexes and conferred highly identifiable shapes to the complexes that facilitated particle selection and orientation for image processing. This study validates bivalent Fabs as new fiducial markers for cryo-EM analysis of small proteins such as HCV E2 and identifies a new target epitope for vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41245.map.gz emd_41245.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41245-v30.xml emd-41245-v30.xml emd-41245.xml emd-41245.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41245_fsc.xml emd_41245_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41245.png emd_41245.png | 164.5 KB | ||
| Filedesc metadata |  emd-41245.cif.gz emd-41245.cif.gz | 7 KB | ||
| Others |  emd_41245_half_map_1.map.gz emd_41245_half_map_1.map.gz emd_41245_half_map_2.map.gz emd_41245_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41245 http://ftp.pdbj.org/pub/emdb/structures/EMD-41245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41245 | HTTPS FTP |
-Related structure data
| Related structure data |  8tgvMC  8tfeC  8tgzC  8thzC  8txqC  8tzyC  8u9yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41245.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41245.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4875 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41245_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41245_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HCV E2-HC84.26 complex
| Entire | Name: HCV E2-HC84.26 complex |
|---|---|
| Components |
|
-Supramolecule #1: HCV E2-HC84.26 complex
| Supramolecule | Name: HCV E2-HC84.26 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 340 KDa |
-Macromolecule #1: envelope glycoprotein E2
| Macromolecule | Name: envelope glycoprotein E2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hepacivirus C Hepacivirus C |
| Molecular weight | Theoretical: 32.28726 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DSTHVTGGTA SHTTRHFASL FSSGASQRVQ LINTNGSWHI NRTALNCNDS LHTGFLAALF YTHKFNASG CPERMAHCRP IDEFAQGWGP ITYAEGHGSD QRPYCWHYAP RQCGTIPASQ VCGPVYCFTP SPVVVGTTDR F GAPTYTWG ...String: METDTLLLWV LLLWVPGSTG DSTHVTGGTA SHTTRHFASL FSSGASQRVQ LINTNGSWHI NRTALNCNDS LHTGFLAALF YTHKFNASG CPERMAHCRP IDEFAQGWGP ITYAEGHGSD QRPYCWHYAP RQCGTIPASQ VCGPVYCFTP SPVVVGTTDR F GAPTYTWG ENETDVLILN NTRPPQGNWF GCTWMNSTGF TKTCGGPPCN IGGVGNNTLT CPTDCFRKHP EATYTKCGSG PW LTPRCLV DYPYRLWHYP CTVNFTIFKV RMYVGGVEHR LNAACNIGHH HHHH UniProtKB: Genome polyprotein |
-Macromolecule #2: HC84.26 Heavy chain
| Macromolecule | Name: HC84.26 Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.304617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DQVQPVQSGA EVKKPGSSVK VSCEASGGTH SNYVITWVRQ APGQGLEWMG GFIPDFRTAM YAQGFQGRV TITADESTSL AYMELTNLRS EDTAVYYCAR GPLSRGYYDY WGPGTLVTVS ASTKGPSVFP LAPSSKSTSG G TAALGCLV ...String: METDTLLLWV LLLWVPGSTG DQVQPVQSGA EVKKPGSSVK VSCEASGGTH SNYVITWVRQ APGQGLEWMG GFIPDFRTAM YAQGFQGRV TITADESTSL AYMELTNLRS EDTAVYYCAR GPLSRGYYDY WGPGTLVTVS ASTKGPSVFP LAPSSKSTSG G TAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PK SCDKTAG WSHPQFEK |
-Macromolecule #3: HC84.26 Light chain
| Macromolecule | Name: HC84.26 Light chain / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.35215 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DSYVLTQPPS VSVAPGQTAS ITCSGDKLGD KYVSWYQQRP GQSPVLVLYQ DSKRPSGIPE RFSGSNSGN TATLTISGTQ AMDEADYYCQ AWDSSALVFG GGTKLTVLRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF Y PREAKVQW ...String: METDTLLLWV LLLWVPGSTG DSYVLTQPPS VSVAPGQTAS ITCSGDKLGD KYVSWYQQRP GQSPVLVLYQ DSKRPSGIPE RFSGSNSGN TATLTISGTQ AMDEADYYCQ AWDSSALVFG GGTKLTVLRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF Y PREAKVQW KVDNALQSGN SQESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl pH 8.0, 100 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9653 / Average exposure time: 3.8 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)