[English] 日本語
 Yorodumi
Yorodumi- EMDB-41232: Cryo-EM structure of the Methanosarcina mazei glutamine synthetas... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Methanosarcina mazei glutamine synthetase (GS) with Met-Sox-P and ADP | |||||||||
 Map data Map data | Mm GS TS map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glutamine synthetase / GS / transition state / MSO / Met-Sox-P / ADP / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine synthetase / glutamine biosynthetic process / glutamine synthetase activity / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Methanosarcina mazei Go1 (archaea) Methanosarcina mazei Go1 (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Schumacher MA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation. Authors: Maria A Schumacher / Raul Salinas / Brady A Travis / Rajiv Ranjan Singh / Nicholas Lent /  Abstract: Glutamine synthetases (GS) play central roles in cellular nitrogen assimilation. Although GS active-site formation requires the oligomerization of just two GS subunits, all GS form large, multi- ...Glutamine synthetases (GS) play central roles in cellular nitrogen assimilation. Although GS active-site formation requires the oligomerization of just two GS subunits, all GS form large, multi-oligomeric machines. Here we describe a structural dissection of the archaeal Methanosarcina mazei (Mm) GS and its regulation. We show that Mm GS forms unstable dodecamers. Strikingly, we show this Mm GS oligomerization property is leveraged for a unique mode of regulation whereby labile Mm GS hexamers are stabilized by binding the nitrogen regulatory protein, GlnK1. Our GS-GlnK1 structure shows that GlnK1 functions as molecular glue to affix GS hexamers together, stabilizing formation of GS active-sites. These data, therefore, reveal the structural basis for a unique form of enzyme regulation by oligomer modulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41232.map.gz emd_41232.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41232-v30.xml emd-41232-v30.xml emd-41232.xml emd-41232.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41232.png emd_41232.png | 178.2 KB | ||
| Filedesc metadata |  emd-41232.cif.gz emd-41232.cif.gz | 5.5 KB | ||
| Others |  emd_41232_half_map_1.map.gz emd_41232_half_map_1.map.gz emd_41232_half_map_2.map.gz emd_41232_half_map_2.map.gz | 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41232 http://ftp.pdbj.org/pub/emdb/structures/EMD-41232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41232 | HTTPS FTP |
-Validation report
| Summary document |  emd_41232_validation.pdf.gz emd_41232_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41232_full_validation.pdf.gz emd_41232_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_41232_validation.xml.gz emd_41232_validation.xml.gz | 12.9 KB | Display | |
| Data in CIF |  emd_41232_validation.cif.gz emd_41232_validation.cif.gz | 15 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41232 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41232 | HTTPS FTP |
-Related structure data
| Related structure data |  8tfkMC  8tfbC  8tfcC  8tgeC  8ufjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41232.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41232.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mm GS TS map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
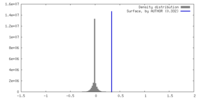
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Mm GS TS half map A
| File | emd_41232_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mm GS TS half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
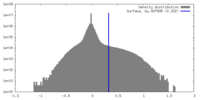
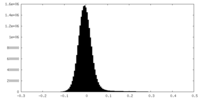
| Density Histograms |
-Half map: Mm GS TS half map B
| File | emd_41232_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mm GS TS half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
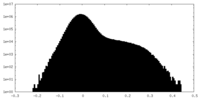
| Density Histograms |
- Sample components
Sample components
-Entire : M. mazei Glutamine Synthetase (GS) transition state complex
| Entire | Name: M. mazei Glutamine Synthetase (GS) transition state complex |
|---|---|
| Components |
|
-Supramolecule #1: M. mazei Glutamine Synthetase (GS) transition state complex
| Supramolecule | Name: M. mazei Glutamine Synthetase (GS) transition state complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Methanosarcina mazei Go1 (archaea) Methanosarcina mazei Go1 (archaea) |
-Macromolecule #1: Glutamine synthetase
| Macromolecule | Name: Glutamine synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: glutamine synthetase |
|---|---|
| Source (natural) | Organism:  Methanosarcina mazei Go1 (archaea) Methanosarcina mazei Go1 (archaea) |
| Molecular weight | Theoretical: 52.827926 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MVQMKKCTTK EDVLEAVKER DVKFIRTQFT DTLGIIKSWA IPAEQLEEAF ENGVMFDGSS IQGFTRIEE SDMKLALDPS TFRILPWRPA TGAVARILGD VYLPDGNPFK GDPRYVLKTA IKEAEKMGFS MNVGPELEFF L FKLDANGN ...String: MGSSHHHHHH SSGLVPRGSH MVQMKKCTTK EDVLEAVKER DVKFIRTQFT DTLGIIKSWA IPAEQLEEAF ENGVMFDGSS IQGFTRIEE SDMKLALDPS TFRILPWRPA TGAVARILGD VYLPDGNPFK GDPRYVLKTA IKEAEKMGFS MNVGPELEFF L FKLDANGN PTTELTDQGG YFDFAPLDRA QDVRRDIDYA LEHMGFQIEA SHHEVAPSQH EIDFRFGDVL CTADNVVTFK YV VKSIAYH KGYYASFMPK PLFGVNGSGM HSNQSLFKDG KNVFYDPDTP TKLSQDAMYY IGGLLKHIRE FTAVTNPVVN SYK RLVPGY EAPVYISWSA QNRSSLIRIP ATRGNGTRIE LRCPDPACNP YLAFALMLRA GLEGIKNKID PGEPTNVNIF HLSD KEREE RGIRSLPADL KEAIDEMKGS KFVKEALGEH VFSHYLCAKE MEWDEYKAVV HPWELSRYLS ML UniProtKB: Glutamine synthetase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 36 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: L-METHIONINE-S-SULFOXIMINE PHOSPHATE
| Macromolecule | Name: L-METHIONINE-S-SULFOXIMINE PHOSPHATE / type: ligand / ID: 3 / Number of copies: 12 / Formula: P3S |
|---|---|
| Molecular weight | Theoretical: 260.205 Da |
| Chemical component information |  ChemComp-P3S: |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 12 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.66 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 164400 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)