[English] 日本語
 Yorodumi
Yorodumi- EMDB-41144: Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPR61 / GPCR / Signaling complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-independent adenylate cyclase-activating G protein-coupled receptor signaling pathway / arrestin family protein binding / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity ...ligand-independent adenylate cyclase-activating G protein-coupled receptor signaling pathway / arrestin family protein binding / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / cellular response to glucagon stimulus / regulation of insulin secretion / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / G protein-coupled receptor activity / bone development / G-protein beta/gamma-subunit complex binding / platelet aggregation / Olfactory Signaling Pathway / cognition / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / sensory perception of smell / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / receptor complex / endosome / endosome membrane / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
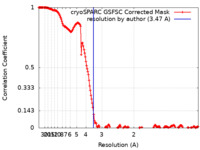
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Lees JA / Dias JM / Han S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism. Authors: Joshua A Lees / João M Dias / Francis Rajamohan / Jean-Philippe Fortin / Rebecca O'Connor / Jimmy X Kong / Emily A G Hughes / Ethan L Fisher / Jamison B Tuttle / Gabrielle Lovett / Bethany ...Authors: Joshua A Lees / João M Dias / Francis Rajamohan / Jean-Philippe Fortin / Rebecca O'Connor / Jimmy X Kong / Emily A G Hughes / Ethan L Fisher / Jamison B Tuttle / Gabrielle Lovett / Bethany L Kormos / Rayomand J Unwalla / Lei Zhang / Anne-Marie Dechert Schmitt / Dahui Zhou / Michael Moran / Kimberly A Stevens / Kimberly F Fennell / Alison E Varghese / Andrew Maxwell / Emmaline E Cote / Yuan Zhang / Seungil Han /  Abstract: GPR61 is an orphan GPCR related to biogenic amine receptors. Its association with phenotypes relating to appetite makes it of interest as a druggable target to treat disorders of metabolism and body ...GPR61 is an orphan GPCR related to biogenic amine receptors. Its association with phenotypes relating to appetite makes it of interest as a druggable target to treat disorders of metabolism and body weight, such as obesity and cachexia. To date, the lack of structural information or a known biological ligand or tool compound has hindered comprehensive efforts to study GPR61 structure and function. Here, we report a structural characterization of GPR61, in both its active-like complex with heterotrimeric G protein and in its inactive state. Moreover, we report the discovery of a potent and selective small-molecule inverse agonist against GPR61 and structural elucidation of its allosteric binding site and mode of action. These findings offer mechanistic insights into an orphan GPCR while providing both a structural framework and tool compound to support further studies of GPR61 function and modulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41144.map.gz emd_41144.map.gz | 450.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41144-v30.xml emd-41144-v30.xml emd-41144.xml emd-41144.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41144_fsc.xml emd_41144_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_41144.png emd_41144.png | 115.1 KB | ||
| Filedesc metadata |  emd-41144.cif.gz emd-41144.cif.gz | 7.3 KB | ||
| Others |  emd_41144_half_map_1.map.gz emd_41144_half_map_1.map.gz emd_41144_half_map_2.map.gz emd_41144_half_map_2.map.gz | 442.9 MB 442.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41144 http://ftp.pdbj.org/pub/emdb/structures/EMD-41144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41144 | HTTPS FTP |
-Validation report
| Summary document |  emd_41144_validation.pdf.gz emd_41144_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41144_full_validation.pdf.gz emd_41144_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_41144_validation.xml.gz emd_41144_validation.xml.gz | 25.4 KB | Display | |
| Data in CIF |  emd_41144_validation.cif.gz emd_41144_validation.cif.gz | 33.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41144 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41144 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41144 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41144 | HTTPS FTP |
-Related structure data
| Related structure data |  8tb0MC  8tb7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41144.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41144.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.59 Å | ||||||||||||||||||||||||||||||||||||
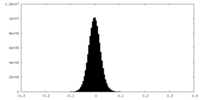
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41144_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
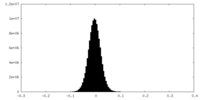
| Density Histograms |
-Half map: #1
| File | emd_41144_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
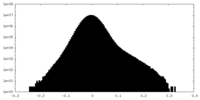
| Density Histograms |
- Sample components
Sample components
-Entire : GPR61-GalphaS-Gbeta1-Ggamma2 complex with single-chain Fv16
| Entire | Name: GPR61-GalphaS-Gbeta1-Ggamma2 complex with single-chain Fv16 |
|---|---|
| Components |
|
-Supramolecule #1: GPR61-GalphaS-Gbeta1-Ggamma2 complex with single-chain Fv16
| Supramolecule | Name: GPR61-GalphaS-Gbeta1-Ggamma2 complex with single-chain Fv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1, #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Single-chain Fv16
| Macromolecule | Name: Single-chain Fv16 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.813047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQG PAAAHHHHHH HH |
-Macromolecule #2: GPR61 fused to dominant negative G alpha S/I N18 chimera
| Macromolecule | Name: GPR61 fused to dominant negative G alpha S/I N18 chimera type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 111.175773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAKGSGAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLLEVLF QGPESSPIPQ SSGNSSTLGR V PQTPGPST ...String: MKTIIALSYI FCLVFADYKD DDDAKGSGAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLLEVLF QGPESSPIPQ SSGNSSTLGR V PQTPGPST ASGVPEVGLR DVASESVALF FMLLLDLTAV AGNAAVMAVI AKTPALRKFV FVFHLCLVDL LAALTLMPLA ML SSSALFD HALFGEVACR LYLFLSVCFV SLAILSVSAI NVERYYYVVH PMRYEVRMTL GLVASVLVGV WVKALAMASV PVL GRVSWE EGAPSVPPGC SLQWSHSAYC QLFVVVFAVL YFLLPLLLIL VVYCSMFRVA RVAAMQHGPL PTWMETPRQR SESL SSRST MVTSSGAPQT TPHRTFGGGK AAVVLLAVGG QFLLCWLPYF SFHLYVALSA QPISTGQVES VVTWIGYFCF TSNPF FYGC LNRQIRGELS KQFVCFFKPA PEEELRLPSR EGSIEENFLQ FLQGTGCPSE SWVSRPLPSP KQEPPAVDFR IPGQIA EET SEFLEQQLTS DIIMSDSYLR PAASPRLESG SSGSSGSSEN LYFQGSGCTL SAEDKAAVER SKMIEKQLQK DKQVYRA TH RLLLLGAGES GKNTIVKQMR ILHVNGFNGE GGEEDPQAAR SNSDGEKATK VQDIKNNLKE AIETIVAAMS NLVPPVEL A NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR ACYERSNEYQ LIDCAQYFLD KIDVIKQADY VPSDQDLLR CRVLTSGIFE TKFQVDKVNF HMFDVGAQRD ERRKWIQCFN DVTAIIFVVA SSSYNMVIRE DNQTNRLQAA LKLFDSIWNN KWLRDTSVI LFLNKQDLLA EKVLAGKSKI EDYFPEFARY TTPEDATPEP GEDPRVTRAK YFIRDEFLRI STASGDGRHY C YPHFTCSV DTENIRRVFN DCRDIIQRMH LRQYELL UniProtKB: G-protein coupled receptor 61, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)