+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RSV preF in complex with Fab 2.4K | |||||||||
 Map data Map data | Sharpened map of of prefusion-stabilized RSV F (PR-DM) complexed with the antigen binding fragment (Fab) from a member of the RSV F public clonotype 2 (2.4K Fab). Sharpened by DeepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / antibody / fusion protein / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated induction of syncytium formation / host cell Golgi membrane / entry receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane Similarity search - Function | |||||||||
| Biological species |  Respiratory syncytial virus A2 / Respiratory syncytial virus A2 /  Homo sapiens (human) / Homo sapiens (human) /  Respiratory syncytial virus Respiratory syncytial virus | |||||||||
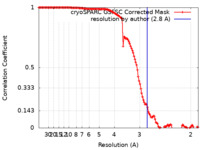
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | McCool RS / McLellan JS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein elicits antibodies targeting a membrane-proximal epitope. Authors: Ryan S McCool / Maryam Musayev / Sabrina M Bush / Alexandrine Derrien-Colemyn / Cory M Acreman / Daniel Wrapp / Tracy J Ruckwardt / Barney S Graham / John R Mascola / Jason S McLellan /  Abstract: Respiratory syncytial virus (RSV) is the leading cause of bronchiolitis and pneumonia in infants, infecting all children by age 5. RSV also causes substantial morbidity and mortality in older adults, ...Respiratory syncytial virus (RSV) is the leading cause of bronchiolitis and pneumonia in infants, infecting all children by age 5. RSV also causes substantial morbidity and mortality in older adults, and a vaccine for older adults based on a prefusion-stabilized form of the viral F glycoprotein was recently approved by the FDA. Here, we investigate a set of antibodies that belong to the same public clonotype and were isolated from individuals vaccinated with a prefusion-stabilized RSV F protein. Our results reveal that these antibodies are highly potent and recognize a previously uncharacterized antigenic site on the prefusion F protein. Vaccination with prefusion RSV F proteins appears to boost the elicitation of these neutralizing antibodies, which are not commonly elicited by natural infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41089.map.gz emd_41089.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41089-v30.xml emd-41089-v30.xml emd-41089.xml emd-41089.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41089_fsc.xml emd_41089_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41089.png emd_41089.png | 58.5 KB | ||
| Masks |  emd_41089_msk_1.map emd_41089_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41089.cif.gz emd-41089.cif.gz | 6.6 KB | ||
| Others |  emd_41089_additional_1.map.gz emd_41089_additional_1.map.gz emd_41089_half_map_1.map.gz emd_41089_half_map_1.map.gz emd_41089_half_map_2.map.gz emd_41089_half_map_2.map.gz | 108.4 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41089 http://ftp.pdbj.org/pub/emdb/structures/EMD-41089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41089 | HTTPS FTP |
-Related structure data
| Related structure data |  8t7aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41089.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41089.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of of prefusion-stabilized RSV F (PR-DM) complexed with the antigen binding fragment (Fab) from a member of the RSV F public clonotype 2 (2.4K Fab). Sharpened by DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
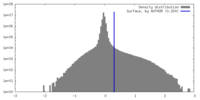
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41089_msk_1.map emd_41089_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map of of prefusion-stabilized RSV F (PR-DM)...
| File | emd_41089_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of of prefusion-stabilized RSV F (PR-DM) complexed with the antigen binding fragment (Fab) from a member of the RSV F public clonotype 2 (2.4K Fab). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of prefusion-stabilized RSV F (PR-DM)...
| File | emd_41089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of prefusion-stabilized RSV F (PR-DM) complexed with the antigen binding fragment (Fab) from a member of the RSV F public clonotype 2 (2.4K Fab). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of prefusion-stabilized RSV F (PR-DM)...
| File | emd_41089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of prefusion-stabilized RSV F (PR-DM) complexed with the antigen binding fragment (Fab) from a member of the RSV F public clonotype 2 (2.4K Fab). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion-stabilized respiratory syncytial virus in complex with ...
| Entire | Name: Prefusion-stabilized respiratory syncytial virus in complex with Fab 2.4K |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion-stabilized respiratory syncytial virus in complex with ...
| Supramolecule | Name: Prefusion-stabilized respiratory syncytial virus in complex with Fab 2.4K type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4 |
|---|
-Supramolecule #2: Prefusion-stabilized respiratory syncytial virus
| Supramolecule | Name: Prefusion-stabilized respiratory syncytial virus / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
-Supramolecule #3: Fab 2.4K
| Supramolecule | Name: Fab 2.4K / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fusion glycoprotein F2
| Macromolecule | Name: Fusion glycoprotein F2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus / Strain: A2 Respiratory syncytial virus / Strain: A2 |
| Molecular weight | Theoretical: 9.498826 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QNITEEFYQS TCSAVSKGYL SALRTGWYTS VITIELSNIK EIKCNGTDAK VKLIKQELDK YKNAVTELQL LMQSTPAANN RARR UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: Fusion glycoprotein F1
| Macromolecule | Name: Fusion glycoprotein F1 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus / Strain: A2 Respiratory syncytial virus / Strain: A2 |
| Molecular weight | Theoretical: 48.295859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FLGFLLGVGS AIASGVAVSK VLHLEGEVNK IKSALLSTNK AVVSLSNGVS VLTSKVLDLK NYIDKQLLPI VNKQSCSIPN IETVIEFQQ KNNRLLEITR EFSVNAGVTT PVSTYMLTNS ELLSLINDMP ITNDQKKLMS NNVQIVRQQS YSIMSIIKEE V LAYVVQLP ...String: FLGFLLGVGS AIASGVAVSK VLHLEGEVNK IKSALLSTNK AVVSLSNGVS VLTSKVLDLK NYIDKQLLPI VNKQSCSIPN IETVIEFQQ KNNRLLEITR EFSVNAGVTT PVSTYMLTNS ELLSLINDMP ITNDQKKLMS NNVQIVRQQS YSIMSIIKEE V LAYVVQLP LYGVIDTPCW KLHTSPLCTT NTKEGSNICL TRTDRGWYCD NAGSVSFFPQ AETCKVQSNR VFCDTMNSLT LP SEVNLCN VDIFNPKYDC KIMTSKTDVS SSVITSLGAI VSCYGKTKCT ASNKNRGIIK TFSNGCDYVS NKGVDTVSVG NTL YYVNKQ EGKSLYVKGE PIINFYDPLV FPSDEFDASI SQVNEKINQS LAFIRKSDEL LGSGYIPEAP RDGQAYVRKD GEWV LLSTF LGRSLEVLFQ GPGHHHHHHH HSAWSHPQFE K UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #3: 2.4K Fab Heavy Chain
| Macromolecule | Name: 2.4K Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.694181 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGGE VKKPGASVKV SCKASGYTFT YYGISWVRQA PGQGLEWMGW ISAYNGNTNY EQKFQGRVTM TTDTSTGTAY MELRSLTSD DTAVYYCARD RIVVVTAANY YGLDVWGQGT TVTVSS |
-Macromolecule #4: 2.4K Fab Light Chain
| Macromolecule | Name: 2.4K Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.447853 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQLTQSPDS LAVSLGERAT INCKSSQSVL YRPNNKNFLA WYQQKPGQPP KLLIYWASTR QSGVPDRFSG SGSGTDFTLT ISSLQAEDV AVYYCQQYHT TPLTFGGGTK VDIK |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)