+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of immature Zika virus | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Zika virus / immature / Cryo-EM / SPA / Homology modeling / prM cleavage / M protein / VIRUS | |||||||||
| Biological species |   Zika virus Zika virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.3 Å | |||||||||
 Authors Authors | Moustafa IM / Hafenstein SL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Npj Viruses / Year: 2023 Journal: Npj Viruses / Year: 2023Title: Zika virus M protein latches and locks the E protein from transitioning to an immature state after prM cleavage. Authors: Sydney A Majowicz / Anoop Narayanan / Ibrahim M Moustafa / Carol M Bator / Susan L Hafenstein / Joyce Jose /  Abstract: During flavivirus maturation, the structural proteins prM (pre-membrane) and E (envelope) undergo extensive low pH-mediated conformational changes, transitioning from spiky trimeric to smooth dimeric ...During flavivirus maturation, the structural proteins prM (pre-membrane) and E (envelope) undergo extensive low pH-mediated conformational changes, transitioning from spiky trimeric to smooth dimeric prM/E heterodimers which allow for furin cleavage of prM into pr and M and forms the irreversible mature conformation of smooth M/E heterodimers. The mechanisms of irreversible conformational changes to E protein following the pr cleavage are not understood. Utilizing cryo-EM structures of immature virus and structure-based mutagenesis of Zika virus, we identified two critical "latching and locking" interactions mediated by M protein residues Arg38 and Trp19, respectively, that stabilize the E protein structure in the smooth mature stage. M protein thus latches and locks the E protein in an irreversible mature structure, preventing premature fusion in the secretory pathway. Our studies provide mechanistic insights into the reversible structural transition of immature trimeric spikes and the irreversible transition of smooth dimeric M/E heterodimers critical for virus infectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41037.map.gz emd_41037.map.gz | 228.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41037-v30.xml emd-41037-v30.xml emd-41037.xml emd-41037.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41037_fsc.xml emd_41037_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41037.png emd_41037.png | 54.6 KB | ||
| Masks |  emd_41037_msk_1.map emd_41037_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41037.cif.gz emd-41037.cif.gz | 5.9 KB | ||
| Others |  emd_41037_half_map_1.map.gz emd_41037_half_map_1.map.gz emd_41037_half_map_2.map.gz emd_41037_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41037 http://ftp.pdbj.org/pub/emdb/structures/EMD-41037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41037 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41037.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41037.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
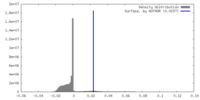
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41037_msk_1.map emd_41037_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
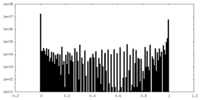
| Density Histograms |
-Half map: half-A map
| File | emd_41037_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
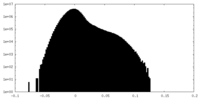
| Density Histograms |
-Half map: half-B map
| File | emd_41037_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
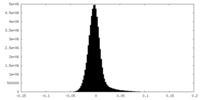
| Density Histograms |
- Sample components
Sample components
-Entire : Zika virus
| Entire | Name:   Zika virus Zika virus |
|---|---|
| Components |
|
-Supramolecule #1: Zika virus
| Supramolecule | Name: Zika virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 64320 / Sci species name: Zika virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: prM/E protein / Diameter: 560.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: E-protein
| Macromolecule | Name: E-protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Sequence | String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQY VCKRTLVDRG WGNGCGLFGK GSLVTCAKFT CSKKMTGKSI QPENLEYRIM LSVHGSQHSG MIVNDTGYET DENRAKVEVT ...String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQY VCKRTLVDRG WGNGCGLFGK GSLVTCAKFT CSKKMTGKSI QPENLEYRIM LSVHGSQHSG MIVNDTGYET DENRAKVEVT PNSPRAEATL GGFGSLGLDC EPRTGLDFSD LYYLTMNNKH WLVHKEWFHD IPLPWHAGAD TGTPHWNNKE ALVEFKDAHA KRQTVVVLGS QEGAVHTALA GALEAEMDGA KGKLFSGHLK CRLKMDKLRL KGVSYSLCTA AFTFTKVPAE TLHGTVTVEV QYAGTDGPCK IPVQMAVDMQ TLTPVGRLIT ANPVITESTE NSKMMLELDP PFGDSYIVIG VGDKKITHHW HRSGSTIGKA FEATVRGAKR MAVLGDTAWD FGSVGGVFNS LGKGIHQIFG AAFKSLFGGM SWFSQILIGT LLVWLGLNTK NGSISLTCLA LGGVMIFLST AVSA |
-Macromolecule #2: prM-protein
| Macromolecule | Name: prM-protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Sequence | String: AEITRRGSAY YMYLDRSDAG KAISFATTLG VNKCHVQIMD LGHMCDATMS YECPMLDEGV EPDDVDCWCN TTSTWVVYGT CHHKKGEARR SRRAVTLPSH STRKLQTRSQ TWLESREYTK HLIKVENWIF RNPGFALVAV AIAWLLGSST SQKVIYLVMI LLIAPAYS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TNE buffer |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K |
| Details | purified immature zika virus at 1.4 mg/ml |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K |
| Specialist optics | Spherical aberration corrector: Microscope was modified with Cs corrector |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1839 / Average exposure time: 90.8 sec. / Average electron dose: 48.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.05 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 59000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Modeller / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)