[English] 日本語
 Yorodumi
Yorodumi- EMDB-41028: Transporter associated with antigen processing (TAP) bound to the... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Transporter associated with antigen processing (TAP) bound to the 9-mer peptide ILKEPVHGV | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / antigen processing / peptide transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationantigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent / tapasin binding / ABC-type peptide antigen transporter activity / ABC-type antigen peptide transporter / TAP complex / ABC-type peptide transporter activity / TAP2 binding / TAP1 binding / peptide antigen transport / MHC class Ib protein binding ...antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent / tapasin binding / ABC-type peptide antigen transporter activity / ABC-type antigen peptide transporter / TAP complex / ABC-type peptide transporter activity / TAP2 binding / TAP1 binding / peptide antigen transport / MHC class Ib protein binding / cytosol to endoplasmic reticulum transport / peptide transport / peptide transmembrane transporter activity / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent / MHC class I protein binding / endoplasmic reticulum-Golgi intermediate compartment membrane / HIV-1 retropepsin / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / T cell mediated cytotoxicity / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / exoribonuclease H activity / response to molecule of bacterial origin / MHC class I peptide loading complex / defense response / antigen processing and presentation of endogenous peptide antigen via MHC class I / ADP binding / DNA integration / viral genome integration into host DNA / positive regulation of T cell mediated cytotoxicity / establishment of integrated proviral latency / RNA-directed DNA polymerase / transmembrane transport / RNA stem-loop binding / centriolar satellite / viral penetration into host nucleus / peptide antigen binding / host multivesicular body / phagocytic vesicle membrane / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / protein transport / host cell / ER-Phagosome pathway / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / adaptive immune response / DNA-directed DNA polymerase activity / nuclear speck / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / endoplasmic reticulum membrane / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / endoplasmic reticulum / protein homodimerization activity / ATP hydrolysis activity / proteolysis / DNA binding / zinc ion binding / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
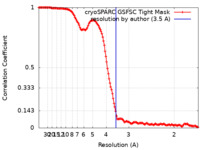
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Lee J / Oldham ML / Chen J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Principles of peptide selection by the transporter associated with antigen processing. Authors: James Lee / Michael L Oldham / Victor Manon / Jue Chen /  Abstract: Our ability to fight pathogens relies on major histocompatibility complex class I (MHC-I) molecules presenting diverse antigens on the surface of diseased cells. The transporter associated with ...Our ability to fight pathogens relies on major histocompatibility complex class I (MHC-I) molecules presenting diverse antigens on the surface of diseased cells. The transporter associated with antigen processing (TAP) transports nearly the entire repertoire of antigenic peptides into the endoplasmic reticulum for MHC-I loading. How TAP transports peptides specific for MHC-I is unclear. In this study, we used cryo-EM to determine a series of structures of human TAP, both in the absence and presence of peptides with various sequences and lengths. The structures revealed that peptides of eight or nine residues in length bind in a similarly extended conformation, despite having little sequence overlap. We also identified two peptide-anchoring pockets on either side of the transmembrane cavity, each engaging one end of a peptide with primarily main chain atoms. Occupation of both pockets results in a global conformational change in TAP, bringing the two halves of the transporter closer together to prime it for isomerization and ATP hydrolysis. Shorter peptides are able to bind to each pocket separately but are not long enough to bridge the cavity to bind to both simultaneously. Mutations that disrupt hydrogen bonds with the N and C termini of peptides almost abolish MHC-I surface expression. Our findings reveal that TAP functions as a molecular caliper that selects peptides according to length rather than sequence, providing antigen diversity for MHC-I presentation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41028.map.gz emd_41028.map.gz | 230.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41028-v30.xml emd-41028-v30.xml emd-41028.xml emd-41028.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41028_fsc.xml emd_41028_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_41028.png emd_41028.png | 40.8 KB | ||
| Filedesc metadata |  emd-41028.cif.gz emd-41028.cif.gz | 6.6 KB | ||
| Others |  emd_41028_additional_1.map.gz emd_41028_additional_1.map.gz emd_41028_half_map_1.map.gz emd_41028_half_map_1.map.gz emd_41028_half_map_2.map.gz emd_41028_half_map_2.map.gz | 122 MB 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41028 http://ftp.pdbj.org/pub/emdb/structures/EMD-41028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41028 | HTTPS FTP |
-Related structure data
| Related structure data |  8t4eMC  8t46C  8t4fC  8t4gC  8t4hC  8t4iC  8t4jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41028.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41028.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.839 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_41028_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41028_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41028_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Transporter associated with antigen processing bound to the 9-mer...
| Entire | Name: Transporter associated with antigen processing bound to the 9-mer peptide ILKEPVHGV |
|---|---|
| Components |
|
-Supramolecule #1: Transporter associated with antigen processing bound to the 9-mer...
| Supramolecule | Name: Transporter associated with antigen processing bound to the 9-mer peptide ILKEPVHGV type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Complex of TAP1 and TAP2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 156 KDa |
-Macromolecule #1: Antigen peptide transporter 1
| Macromolecule | Name: Antigen peptide transporter 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.034289 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASSRCPAPR GCRCLPGASL AWLGTVLLLL ADWVLLRTAL PRIFSLLVPT ALPLLRVWAV GLSRWAVLWL GACGVLRATV GSKSENAGA QGWLAALKPL AAALGLALPG LALFRELISW GAPGSADSTR LLHWGSHPTA FVVSYAAALP AAALWHKLGS L WVPGGQGG ...String: MASSRCPAPR GCRCLPGASL AWLGTVLLLL ADWVLLRTAL PRIFSLLVPT ALPLLRVWAV GLSRWAVLWL GACGVLRATV GSKSENAGA QGWLAALKPL AAALGLALPG LALFRELISW GAPGSADSTR LLHWGSHPTA FVVSYAAALP AAALWHKLGS L WVPGGQGG SGNPVRRLLG CLGSETRRLS LFLVLVVLSS LGEMAIPFFT GRLTDWILQD GSADTFTRNL TLMSILTIAS AV LEFVGDG IYNNTMGHVH SHLQGEVFGA VLRQETEFFQ QNQTGNIMSR VTEDTSTLSD SLSENLSLFL WYLVRGLCLL GIM LWGSVS LTMVTLITLP LLFLLPKKVG KWYQLLEVQV RESLAKSSQV AIEALSAMPT VRSFANEEGE AQKFREKLQE IKTL NQKEA VAYAVNSWTT SISGMLLKVG ILYIGGQLVT SGAVSSGNLV TFVLYQMQFT QAVEVLLSIY PRVQKAVGSS EKIFE YLDR TPRCPPSGLL TPLHLEGLVQ FQDVSFAYPN RPDVLVLQGL TFTLRPGEVT ALVGPNGSGK STVAALLQNL YQPTGG QLL LDGKPLPQYE HRYLHRQVAA VGQEPQVFGR SLQENIAYGL TQKPTMEEIT AAAVKSGAHS FISGLPQGYD TEVDEAG SQ LSGGQRQAVA LARALIRKPC VLILDDATSA LDANSQLQVE QLLYESPERY SRSVLLITQH LSLVEQADHI LFLEGGAI R EGGTHQQLME KKGCYWAMVQ APADAPE UniProtKB: Antigen peptide transporter 1 |
-Macromolecule #2: Antigen peptide transporter 2
| Macromolecule | Name: Antigen peptide transporter 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.736508 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRLPDLRPWT SLLLVDAALL WLLQGPLGTL LPQGLPGLWL EGTLRLGGLW GLLKLRGLLG FVGTLLLPLC LATPLTVSLR ALVAGASRA PPARVASAPW SWLLVGYGAA GLSWSLWAVL SPPGAQEKEQ DQVNNKVLMW RLLKLSRPDL PLLVAAFFFL V LAVLGETL ...String: MRLPDLRPWT SLLLVDAALL WLLQGPLGTL LPQGLPGLWL EGTLRLGGLW GLLKLRGLLG FVGTLLLPLC LATPLTVSLR ALVAGASRA PPARVASAPW SWLLVGYGAA GLSWSLWAVL SPPGAQEKEQ DQVNNKVLMW RLLKLSRPDL PLLVAAFFFL V LAVLGETL IPHYSGRVID ILGGDFDPHA FASAIFFMCL FSFGSSLSAG CRGGCFTYTM SRINLRIREQ LFSSLLRQDL GF FQETKTG ELNSRLSSDT TLMSNWLPLN ANVLLRSLVK VVGLYGFMLS ISPRLTLLSL LHMPFTIAAE KVYNTRHQEV LRE IQDAVA RAGQVVREAV GGLQTVRSFG AEEHEVCRYK EALEQCRQLY WRRDLERALY LLVRRVLHLG VQMLMLSCGL QQMQ DGELT QGSLLSFMIY QESVGSYVQT LVYIYGDMLS NVGAAEKVFS YMDRQPNLPS PGTLAPTTLQ GVVKFQDVSF AYPNR PDRP VLKGLTFTLR PGEVTALVGP NGSGKSTVAA LLQNLYQPTG GQVLLDEKPI SQYEHCYLHS QVVSVGQEPV LFSGSV RNN IAYGLQSCED DKVMAAAQAA HADDFIQEME HGIYTDVGEK GSQLAAGQKQ RLAIARALVR DPRVLILDEA TSALDVQ CE QALQDWNSRG DRTVLVIAHR LQTVQRAHQI LVLQEGKLQK LAQL UniProtKB: Antigen peptide transporter 2 |
-Macromolecule #3: Transframe peptide
| Macromolecule | Name: Transframe peptide / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 993.199 Da |
| Sequence | String: ILKEPVHGV UniProtKB: Gag-Pol polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)