[English] 日本語
 Yorodumi
Yorodumi- EMDB-40965: AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide; focused re... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide; focused refinement 3 | |||||||||
 Map data Map data | Sharpened map from cryoSPARC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Clathrin-mediated endocytosis / peripheral membrane protein / ENDOCYTOSIS | |||||||||
| Biological species |  | |||||||||
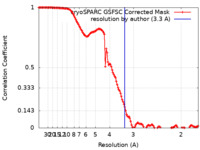
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Baker RW / Sarsam R / Cannon KS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Lipid nanodiscs as a template for high-resolution cryo-EM structures of peripheral membrane proteins. Authors: Kevin S Cannon / Reta D Sarsam / Tanita Tedamrongwanish / Kevin Zhang / Richard W Baker /  Abstract: Peripheral membrane proteins are ubiquitous throughout cell biology and are required for a variety of cellular processes such as signal transduction, membrane trafficking, and autophagy. Transient ...Peripheral membrane proteins are ubiquitous throughout cell biology and are required for a variety of cellular processes such as signal transduction, membrane trafficking, and autophagy. Transient binding to the membrane has a profound impact on protein function, serving to induce conformational changes and alter biochemical and biophysical parameters by increasing the local concentration of factors and restricting diffusion to two dimensions. Despite the centrality of the membrane in serving as a template for cell biology, there are few reported high-resolution structures of peripheral membrane proteins bound to the membrane. We analyzed the utility of lipid nanodiscs to serve as a template for cryo-EM analysis of peripheral membrane proteins. We tested a variety of nanodiscs and we report a 3.3 Å structure of the AP2 clathrin adaptor complex bound to a 17-nm nanodisc, with sufficient resolution to visualize a bound lipid head group. Our data demonstrate that lipid nanodiscs are amenable to high-resolution structure determination of peripheral membrane proteins and provide a framework for extending this analysis to other systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40965.map.gz emd_40965.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40965-v30.xml emd-40965-v30.xml emd-40965.xml emd-40965.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40965_fsc.xml emd_40965_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40965.png emd_40965.png | 44.8 KB | ||
| Masks |  emd_40965_msk_1.map emd_40965_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_40965_additional_1.map.gz emd_40965_additional_1.map.gz emd_40965_additional_2.map.gz emd_40965_additional_2.map.gz emd_40965_additional_3.map.gz emd_40965_additional_3.map.gz emd_40965_additional_4.map.gz emd_40965_additional_4.map.gz emd_40965_half_map_1.map.gz emd_40965_half_map_1.map.gz emd_40965_half_map_2.map.gz emd_40965_half_map_2.map.gz | 188.8 MB 108.1 MB 1.8 MB 1.9 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40965 http://ftp.pdbj.org/pub/emdb/structures/EMD-40965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40965 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40965.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40965.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from cryoSPARC | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40965_msk_1.map emd_40965_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
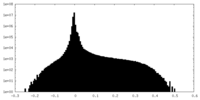
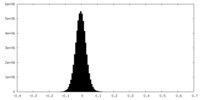
| Density Histograms |
-Additional map: Sharpened map from deepEMhancer
| File | emd_40965_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
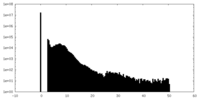
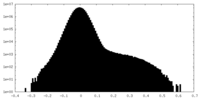
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_40965_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
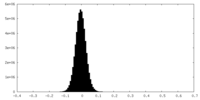
| Density Histograms |
-Additional map: LocRes file for coloring by local resolution
| File | emd_40965_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocRes file for coloring by local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
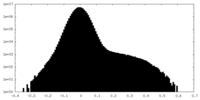
| Density Histograms |
-Additional map: Map filtered by local resolution
| File | emd_40965_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map filtered by local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
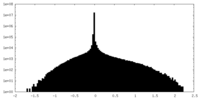
| Density Histograms |
-Half map: Half map 2
| File | emd_40965_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_40965_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide
| Entire | Name: AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide |
|---|---|
| Components |
|
-Supramolecule #1: AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide
| Supramolecule | Name: AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Nanodiscs were assembled with a lipid mixture containing 75 mol% DOPC, 15 mol% DOPS, 10 mol% PIP2. Complex was formed by co-elution via gel filtration chromatography. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM HEPES pH 7.4, 100 mM NaCl |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Two applications of sample.. |
| Details | Nanodiscs were assembled with a lipid mixture containing 75 mol% DOPC, 15 mol% DOPS, 10 mol% PIP2. Complex was formed by co-elution via gel filtration chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 45000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 45000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)