[English] 日本語
 Yorodumi
Yorodumi- EMDB-40946: Cryo-EM of the GDP-bound human dynamin (full-length) polymer asse... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of the GDP-bound human dynamin (full-length) polymer assembled on the membrane in the super constricted state (full helix) | ||||||||||||
 Map data Map data | GDP-bound human dynamin (full-length) polymer assembled on the membrane in the super constricted state (full helix) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | dynamin / membrane / fission / lipid / tubule / scission / endocytosis / HYDROLASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin coat assembly involved in endocytosis / vesicle scission / synaptic vesicle budding from presynaptic endocytic zone membrane / dynamin GTPase / chromaffin granule / regulation of vesicle size / Retrograde neurotrophin signalling / Toll Like Receptor 4 (TLR4) Cascade / Formation of annular gap junctions / endosome organization ...clathrin coat assembly involved in endocytosis / vesicle scission / synaptic vesicle budding from presynaptic endocytic zone membrane / dynamin GTPase / chromaffin granule / regulation of vesicle size / Retrograde neurotrophin signalling / Toll Like Receptor 4 (TLR4) Cascade / Formation of annular gap junctions / endosome organization / Gap junction degradation / phosphatidylinositol-3,4,5-trisphosphate binding / Recycling pathway of L1 / EPH-ephrin mediated repulsion of cells / endocytic vesicle / phosphatidylinositol-4,5-bisphosphate binding / clathrin-coated pit / MHC class II antigen presentation / receptor-mediated endocytosis / cell projection / protein homooligomerization / receptor internalization / endocytosis / GDP binding / presynapse / Clathrin-mediated endocytosis / protein homotetramerization / microtubule binding / microtubule / GTPase activity / synapse / protein kinase binding / GTP binding / protein homodimerization activity / RNA binding / extracellular exosome / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
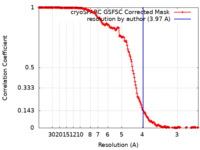
| Method | helical reconstruction / cryo EM / Resolution: 3.97 Å | ||||||||||||
 Authors Authors | Jimah JR / Canagarajah BJ / Hinshaw JE | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Dev Cell / Year: 2024 Journal: Dev Cell / Year: 2024Title: Cryo-EM structures of membrane-bound dynamin in a post-hydrolysis state primed for membrane fission. Authors: John R Jimah / Nidhi Kundu / Abigail E Stanton / Kem A Sochacki / Bertram Canagarajah / Lieza Chan / Marie-Paule Strub / Huaibin Wang / Justin W Taraska / Jenny E Hinshaw /  Abstract: Dynamin assembles as a helical polymer at the neck of budding endocytic vesicles, constricting the underlying membrane as it progresses through the GTPase cycle to sever vesicles from the ...Dynamin assembles as a helical polymer at the neck of budding endocytic vesicles, constricting the underlying membrane as it progresses through the GTPase cycle to sever vesicles from the plasma membrane. Although atomic models of the dynamin helical polymer bound to guanosine triphosphate (GTP) analogs define earlier stages of membrane constriction, there are no atomic models of the assembled state post-GTP hydrolysis. Here, we used cryo-EM methods to determine atomic structures of the dynamin helical polymer assembled on lipid tubules, akin to necks of budding endocytic vesicles, in a guanosine diphosphate (GDP)-bound, super-constricted state. In this state, dynamin is assembled as a 2-start helix with an inner lumen of 3.4 nm, primed for spontaneous fission. Additionally, by cryo-electron tomography, we trapped dynamin helical assemblies within HeLa cells using the GTPase-defective dynamin K44A mutant and observed diverse dynamin helices, demonstrating that dynamin can accommodate a range of assembled complexes in cells that likely precede membrane fission. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40946.map.gz emd_40946.map.gz | 51.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40946-v30.xml emd-40946-v30.xml emd-40946.xml emd-40946.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40946_fsc.xml emd_40946_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40946.png emd_40946.png | 113.7 KB | ||
| Masks |  emd_40946_msk_1.map emd_40946_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40946.cif.gz emd-40946.cif.gz | 6.8 KB | ||
| Others |  emd_40946_half_map_1.map.gz emd_40946_half_map_1.map.gz emd_40946_half_map_2.map.gz emd_40946_half_map_2.map.gz | 442 MB 442 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40946 http://ftp.pdbj.org/pub/emdb/structures/EMD-40946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40946 | HTTPS FTP |
-Related structure data
| Related structure data |  8t0rMC  8sxzC  8sz4C  8sz7C  8sz8C  8t0kC  8tymC  8tynC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40946.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40946.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GDP-bound human dynamin (full-length) polymer assembled on the membrane in the super constricted state (full helix) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.289 Å | ||||||||||||||||||||||||||||||||||||
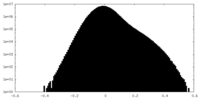
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40946_msk_1.map emd_40946_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
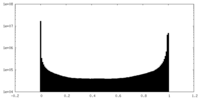
| Density Histograms |
-Half map: Half Map 1
| File | emd_40946_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
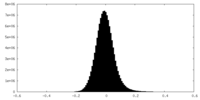
| Density Histograms |
-Half map: Half Map 2
| File | emd_40946_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
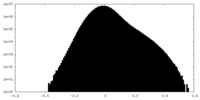
| Density Histograms |
- Sample components
Sample components
-Entire : GDP-bound human dynamin (full-length) helical polymer assembled o...
| Entire | Name: GDP-bound human dynamin (full-length) helical polymer assembled on the membrane (full helix) |
|---|---|
| Components |
|
-Supramolecule #1: GDP-bound human dynamin (full-length) helical polymer assembled o...
| Supramolecule | Name: GDP-bound human dynamin (full-length) helical polymer assembled on the membrane (full helix) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.7 kDa/nm |
-Macromolecule #1: Dynamin-1
| Macromolecule | Name: Dynamin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: dynamin GTPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.825742 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAHHHHHHTG MGNRGMEDLI PLVNRLQDAF SAIGQNADLD LPQIAVVGGQ SAGASSVLEN FVGRDFLPRG SGIVTRRPLV LQLVNATTE YAEFLHCKGK KFTDFEEVRL EIEAETDRVT GTNKGISPVP INLRVYSPHV LNLTLVDLPG MTKVPVGDQP P DIEFQIRD ...String: MAHHHHHHTG MGNRGMEDLI PLVNRLQDAF SAIGQNADLD LPQIAVVGGQ SAGASSVLEN FVGRDFLPRG SGIVTRRPLV LQLVNATTE YAEFLHCKGK KFTDFEEVRL EIEAETDRVT GTNKGISPVP INLRVYSPHV LNLTLVDLPG MTKVPVGDQP P DIEFQIRD MLMQFVTKEN CLILAVSPAN SDLANSDALK VAKEVDPQGQ RTIGVITKLD LMDEGTDARD VLENKLLPLR RG YIGVVNR SQKDIDGKKD ITAALAAERK FFLSHPSYRH LADRMGTPYL QKVLNQQLTN HIRDTLPGLR NKLQSQLLSI EKE VEEYKN FRPDDPARKT KALLQMVQQF AVDFEKRIEG SGDQIDTYEL SGGARINRIF HERFPFELVK MEFDEKELRR EISY AIKNI HGIRTGLFTP DMAFETIVKK QVKKIREPCL KCVDMVISEL ISTVRQCTKK LQQYPRLREE MERIVTTHIR EREGR TKEQ VMLLIDIELA YMNTNHEDFI GFANAQQRSN QMNKKKTSGN QDEILVIRKG WLTINNIGIM KGGSKEYWFV LTAENL SWY KDDEEKEKKY MLSVDNLKLR DVEKGFMSSK HIFALFNTEQ RNVYKDYRQL ELACETQEEV DSWKASFLRA GVYPERV GD KEKASETEEN GSDSFMHSMD PQLERQVETI RNLVDSYMAI VNKTVRDLMP KTIMHLMINN TKEFIFSELL ANLYSCGD Q NTLMEESAEQ AQRRDEMLRM YHALKEALSI IGNINTTTVS TPMPPPVDDS WLQVQSVPAG RRSPTSSPTP QRRAPAVPP ARPGSRGPAP GPPPAGSALG GAPPVPSRPG ASPDPFGPPP QVPSRPNRAP PGVPSRSGQA SPSRPESPRP PFDLGT UniProtKB: Dynamin-1 |
-Macromolecule #2: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)