[English] 日本語
 Yorodumi
Yorodumi- EMDB-40890: Human heavy chain apoferritin prepared with axisymmetric blotting. -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human heavy chain apoferritin prepared with axisymmetric blotting. | |||||||||
 Map data Map data | Human heavy chain apoferritin prepared with axisymmetric blotting | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Iron binding / METAL BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
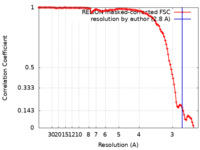
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Glaeser RM / Han BG / Avila-Sakar A / Remis JP | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Curr Opin Struct Biol / Year: 2023 Journal: Curr Opin Struct Biol / Year: 2023Title: Challenges in making ideal cryo-EM samples. Authors: Bong-Gyoon Han / Agustin Avila-Sakar / Jonathan Remis / Robert M Glaeser /  Abstract: Recognizing that interaction with the air-water interface (AWI) is a major challenge for cryo-EM, we first review current approaches designed to avoid it. Of these, immobilizing particles on affinity ...Recognizing that interaction with the air-water interface (AWI) is a major challenge for cryo-EM, we first review current approaches designed to avoid it. Of these, immobilizing particles on affinity grids is arguably the most promising. In addition, we review efforts to gain more reliable control of the sample thicknesses, not the least important reason being to prevent immobilized particles from coming in contact with the AWI of the remaining buffer. It is emphasized that avoiding such a contact is as important for cryo-ET as for single-particle cryo-EM. Finally, looking to the future, it is proposed that immobilized samples might be used to perform time-resolved biochemical experiments directly on EM grids rather than just in test tubes or cuvettes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40890.map.gz emd_40890.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40890-v30.xml emd-40890-v30.xml emd-40890.xml emd-40890.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40890_fsc.xml emd_40890_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_40890.png emd_40890.png | 92.9 KB | ||
| Others |  emd_40890_half_map_1.map.gz emd_40890_half_map_1.map.gz emd_40890_half_map_2.map.gz emd_40890_half_map_2.map.gz | 93.3 MB 93.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40890 http://ftp.pdbj.org/pub/emdb/structures/EMD-40890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40890 | HTTPS FTP |
-Validation report
| Summary document |  emd_40890_validation.pdf.gz emd_40890_validation.pdf.gz | 558.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40890_full_validation.pdf.gz emd_40890_full_validation.pdf.gz | 558.2 KB | Display | |
| Data in XML |  emd_40890_validation.xml.gz emd_40890_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_40890_validation.cif.gz emd_40890_validation.cif.gz | 24.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40890 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40890 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human heavy chain apoferritin prepared with axisymmetric blotting | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40890_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
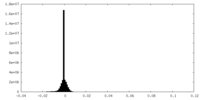
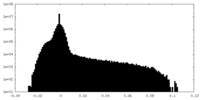
| Density Histograms |
-Half map: Half Map 2
| File | emd_40890_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
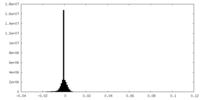
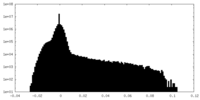
| Density Histograms |
- Sample components
Sample components
-Entire : ApoFerritin
| Entire | Name: ApoFerritin |
|---|---|
| Components |
|
-Supramolecule #1: ApoFerritin
| Supramolecule | Name: ApoFerritin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 470 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)