+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of LINE-1 ORF2p with template:primer hybrid | |||||||||
 Map data Map data | Main map of LINE-1 ORF2p post-processed with deepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | reverse transcriptase / LINE-1 / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleic acid metabolic process / retrotransposition / type II site-specific deoxyribonuclease activity / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / RNA-directed DNA polymerase / RNA-directed DNA polymerase activity / DNA recombination / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
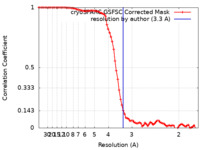
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | van Eeuwen T / Taylor MS / Rout MP | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structures, functions and adaptations of the human LINE-1 ORF2 protein. Authors: Eric T Baldwin / Trevor van Eeuwen / David Hoyos / Arthur Zalevsky / Egor P Tchesnokov / Roberto Sánchez / Bryant D Miller / Luciano H Di Stefano / Francesc Xavier Ruiz / Matthew Hancock / ...Authors: Eric T Baldwin / Trevor van Eeuwen / David Hoyos / Arthur Zalevsky / Egor P Tchesnokov / Roberto Sánchez / Bryant D Miller / Luciano H Di Stefano / Francesc Xavier Ruiz / Matthew Hancock / Esin Işik / Carlos Mendez-Dorantes / Thomas Walpole / Charles Nichols / Paul Wan / Kirsi Riento / Rowan Halls-Kass / Martin Augustin / Alfred Lammens / Anja Jestel / Paula Upla / Kera Xibinaku / Samantha Congreve / Maximiliaan Hennink / Kacper B Rogala / Anna M Schneider / Jennifer E Fairman / Shawn M Christensen / Brian Desrosiers / Gregory S Bisacchi / Oliver L Saunders / Nafeeza Hafeez / Wenyan Miao / Rosana Kapeller / Dennis M Zaller / Andrej Sali / Oliver Weichenrieder / Kathleen H Burns / Matthias Götte / Michael P Rout / Eddy Arnold / Benjamin D Greenbaum / Donna L Romero / John LaCava / Martin S Taylor /      Abstract: The LINE-1 (L1) retrotransposon is an ancient genetic parasite that has written around one-third of the human genome through a 'copy and paste' mechanism catalysed by its multifunctional enzyme, open ...The LINE-1 (L1) retrotransposon is an ancient genetic parasite that has written around one-third of the human genome through a 'copy and paste' mechanism catalysed by its multifunctional enzyme, open reading frame 2 protein (ORF2p). ORF2p reverse transcriptase (RT) and endonuclease activities have been implicated in the pathophysiology of cancer, autoimmunity and ageing, making ORF2p a potential therapeutic target. However, a lack of structural and mechanistic knowledge has hampered efforts to rationally exploit it. We report structures of the human ORF2p 'core' (residues 238-1061, including the RT domain) by X-ray crystallography and cryo-electron microscopy in several conformational states. Our analyses identified two previously undescribed folded domains, extensive contacts to RNA templates and associated adaptations that contribute to unique aspects of the L1 replication cycle. Computed integrative structural models of full-length ORF2p show a dynamic closed-ring conformation that appears to open during retrotransposition. We characterize ORF2p RT inhibition and reveal its underlying structural basis. Imaging and biochemistry show that non-canonical cytosolic ORF2p RT activity can produce RNA:DNA hybrids, activating innate immune signalling through cGAS/STING and resulting in interferon production. In contrast to retroviral RTs, L1 RT is efficiently primed by short RNAs and hairpins, which probably explains cytosolic priming. Other biochemical activities including processivity, DNA-directed polymerization, non-templated base addition and template switching together allow us to propose a revised L1 insertion model. Finally, our evolutionary analysis demonstrates structural conservation between ORF2p and other RNA- and DNA-dependent polymerases. We therefore provide key mechanistic insights into L1 polymerization and insertion, shed light on the evolutionary history of L1 and enable rational drug development targeting L1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40858.map.gz emd_40858.map.gz | 46.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40858-v30.xml emd-40858-v30.xml emd-40858.xml emd-40858.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40858_fsc.xml emd_40858_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40858.png emd_40858.png | 48.5 KB | ||
| Masks |  emd_40858_msk_1.map emd_40858_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40858.cif.gz emd-40858.cif.gz | 8.1 KB | ||
| Others |  emd_40858_additional_1.map.gz emd_40858_additional_1.map.gz emd_40858_half_map_1.map.gz emd_40858_half_map_1.map.gz emd_40858_half_map_2.map.gz emd_40858_half_map_2.map.gz | 27.2 MB 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40858 http://ftp.pdbj.org/pub/emdb/structures/EMD-40858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40858 | HTTPS FTP |
-Validation report
| Summary document |  emd_40858_validation.pdf.gz emd_40858_validation.pdf.gz | 677.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40858_full_validation.pdf.gz emd_40858_full_validation.pdf.gz | 677 KB | Display | |
| Data in XML |  emd_40858_validation.xml.gz emd_40858_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_40858_validation.cif.gz emd_40858_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40858 | HTTPS FTP |
-Related structure data
| Related structure data |  8sxtMC  8c8jC  8sxuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40858.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40858.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map of LINE-1 ORF2p post-processed with deepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40858_msk_1.map emd_40858_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Main map of LINE-1 ORF2p post-processed with global...
| File | emd_40858_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map of LINE-1 ORF2p post-processed with global B factor sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Long Interspersed Nuclear Element (LINE)-1 ORF2 protein
| Entire | Name: Long Interspersed Nuclear Element (LINE)-1 ORF2 protein |
|---|---|
| Components |
|
-Supramolecule #1: Long Interspersed Nuclear Element (LINE)-1 ORF2 protein
| Supramolecule | Name: Long Interspersed Nuclear Element (LINE)-1 ORF2 protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DNA primer
| Macromolecule | Name: DNA primer / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.975611 KDa |
| Sequence | String: (DG)(DC)(DG)(DA)(DA)(DA)(DA)(DT)(DT)(DT) (DC)(DG)(DC) |
-Macromolecule #2: RNA template
| Macromolecule | Name: RNA template / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.444269 KDa |
| Sequence | String: GUGAGCGAAA UUUUCGC |
-Macromolecule #3: LINE-1 retrotransposable element ORF2 protein
| Macromolecule | Name: LINE-1 retrotransposable element ORF2 protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 149.252344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTGSNSHITI LTLNVNGLNS PIKRHRLASW IKSQDPSVCC IQETHLTCRD THRLKIKGWR KIYQANGKQK KAGVAILVSD KTDFKPTKI KRDKEGHYIM VKGSIQQEEL TILNIYAPNT GAPRFIKQVL SDLQRDLDSH TLIMGDFNTP LSILDRSTRQ K VNKDTQEL ...String: MTGSNSHITI LTLNVNGLNS PIKRHRLASW IKSQDPSVCC IQETHLTCRD THRLKIKGWR KIYQANGKQK KAGVAILVSD KTDFKPTKI KRDKEGHYIM VKGSIQQEEL TILNIYAPNT GAPRFIKQVL SDLQRDLDSH TLIMGDFNTP LSILDRSTRQ K VNKDTQEL NSALHQTDLI DIYRTLHPKS TEYTFFSAPH HTYSKIDHIV GSKALLSKCK RTEIITNYLS DHSAIKLELR IK NLTQSRS TTWKLNNLLL NDYWVHNEMK AEIKMFFETN ENKDTTYQNL WDAFKAVCRG KFIALNAYKR KQERSKIDTL TSQ LKELEK QEQTHSKASR RQEITKIRAE LKEIETQKTL QKINESRSWF FERINKIDRP LARLIKKKRE KNQIDTIKND KGDI TTDPT EIQTTIREYY KHLYANKLEN LEEMDTFLDT YTLPRLNQEE VESLNRPITG SEIVAIINSL PTKKSPGPDG FTAEF YQRY KEELVPFLLK LFQSIEKEGI LPNSFYEASI ILIPKPGRDT TKKENFRPIS LMNIDAKILN KILANRIQQH IKKLIH HDQ VGFIPGMQGW FNIRKSINVI QHINRAKDKN HVIISIDAEK AFDKIQQPFM LKTLNKLGID GMYLKIIRAI YDKPTAN II LNGQKLEAFP LKTGTRQGCP LSPLLFNIVL EVLARAIRQE KEIKGIQLGK EEVKLSLFAD DMIVYLENPI VSAQNLLK L ISNFSKVSGY KINVQKSQAF LYNNNRQTES QIMGELPFTI ASKRIKYLGI QLTRDVKDLF KENYKPLLKE IKEDTNKWK NIPCSWVGRI NIVKMAILPK VIYRFNAIPI KLPMTFFTEL EKTTLKFIWN QKRARIAKSI LSQKNKAGGI TLPDFKLYYK ATVTKTAWY WYQNRDIDQW NRTEPSEIMP HIYNYLIFDK PEKNKQWGKD SLLNKWCWEN WLAICRKLKL DPFLTPYTKI N SRWIKDLN VKPKTIKTLE ENLGITIQDI GVGKDFMSKT PKAMATKDKI DKWDLIKLKS FCTAKETTIR VNRQPTTWEK IF ATYSSDK GLISRIYNEL KQIYKKKTNN PIKKWAKDMN RHFSKEDIYA AKKHMKKCSS SLAIREMQIK TTMRYHLTPV RMA IIKKSG NNRCWRGCGE IGTLVHCWWD CKLVQPLWKS VWRFLRDLEL EIPFDPAIPL LGIYPKDYKS CCYKDTCTRM FIAA LFTIA KTWNQPNCPT MIDWIKKMWH IYTMEYYAAI KNDEFISFVG TWMKLETIIL SKLSQEQKTK HRIFSLIGGN UniProtKB: LINE-1 retrotransposable element ORF2 protein |
-Macromolecule #4: THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: TTP |
|---|---|
| Molecular weight | Theoretical: 482.168 Da |
| Chemical component information |  ChemComp-TTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: 20mM HEPES pH 7.6, 150mM NaCl, 2mM MgOAc, 2mM DTT, 2mM dTTP | |||||||||||||||
| Grid | Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: Pelco easy glow | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC / Details: Manually blotted from the back. | |||||||||||||||
| Details | Sample was monodisperse though unstable |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 11270 / Average electron dose: 54.0 e/Å2 Details: Super-resolution images collected in dose-fractionation mode over 54 frames with a dose per frame of 1.08 e/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)