[English] 日本語
 Yorodumi
Yorodumi- EMDB-40786: Structural basis and functional roles for Toll-like receptor bind... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis and functional roles for Toll-like receptor binding to Latrophilin adhesion-GPCR in embryo development | |||||||||
 Map data Map data | Postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | synapse / C. elegans / ADGR / GPCR / CELL ADHESION | |||||||||
| Biological species |  | |||||||||
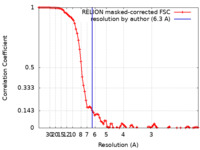
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Li J / Rosas GC / Arac D / Ozkan E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis and functional roles for Toll-like receptor binding to Latrophilin in C. elegans development. Authors: Gabriel Carmona-Rosas / Jingxian Li / Jayson J Smith / Wioletta I Nawrocka / Shouqiang Cheng / Elana E Baltrusaitis / Minglei Zhao / Demet Araç / Paschalis Kratsios / Engin Özkan /  Abstract: Latrophilins are conserved adhesion-type G-protein-coupled receptors associated with embryonic defects and lethality. However, their mechanistic roles and ligands in embryogenesis remain unknown. ...Latrophilins are conserved adhesion-type G-protein-coupled receptors associated with embryonic defects and lethality. However, their mechanistic roles and ligands in embryogenesis remain unknown. Here, we identified TOL-1, the sole Toll-like receptor in Caenorhabditis elegans, as a ligand for the C. elegans latrophilin, LAT-1. The extracellular lectin domain of LAT-1 directly binds to the second leucine-rich repeat domain of TOL-1. The crystal structure and cryo-electron microscopy density map of the LAT-1-TOL-1 extracellular region complex reveal a one-to-one lectin domain interaction with the convex face of a leucine-rich repeat domain. In C. elegans, endogenous mRNA and protein localization analyses showed mutually exclusive sites of expression, suggesting that in vivo LAT-1-TOL-1 interactions mostly occur in trans. Mutagenesis of key interface residues that disrupt the LAT-1-TOL-1 interaction led to partial lethality and malformed embryos. Thus, TOL-1 binding to LAT-1 represents a receptor-ligand axis essential for animal development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40786.map.gz emd_40786.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40786-v30.xml emd-40786-v30.xml emd-40786.xml emd-40786.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40786_fsc.xml emd_40786_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40786.png emd_40786.png | 50.9 KB | ||
| Filedesc metadata |  emd-40786.cif.gz emd-40786.cif.gz | 6.1 KB | ||
| Others |  emd_40786_half_map_1.map.gz emd_40786_half_map_1.map.gz emd_40786_half_map_2.map.gz emd_40786_half_map_2.map.gz | 49.7 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40786 http://ftp.pdbj.org/pub/emdb/structures/EMD-40786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40786 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40786.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40786.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Halp map 1
| File | emd_40786_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halp map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_40786_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The binary complex of C. elegans TOL-1 with C. elegans LAT-1 extr...
| Entire | Name: The binary complex of C. elegans TOL-1 with C. elegans LAT-1 extracellular domains |
|---|---|
| Components |
|
-Supramolecule #1: The binary complex of C. elegans TOL-1 with C. elegans LAT-1 extr...
| Supramolecule | Name: The binary complex of C. elegans TOL-1 with C. elegans LAT-1 extracellular domains type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The Binary complex includes C.elegans Toll-like receptor 1 (Tol-1) and C. elegans Latrophilin 1 (LAT-1). |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 172 KDa |
-Macromolecule #1: Toll-like receptor 1/TOL-1
| Macromolecule | Name: Toll-like receptor 1/TOL-1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ADPHHHHHHG SGLNDIFEAQ KIEWHEADPG YTDECPKFCK CAPDPVQPTS KLLLCDYSSK NTTITPIASS NYDQVANIRS LFISCDNNNF QFPDAYFKSL TALHHLRIVG CETTHFSVKL FEDLAALRRL ELDQISTAST SFEMTEDVLM PLARLEKFSL TRSRNIELPQ ...String: ADPHHHHHHG SGLNDIFEAQ KIEWHEADPG YTDECPKFCK CAPDPVQPTS KLLLCDYSSK NTTITPIASS NYDQVANIRS LFISCDNNNF QFPDAYFKSL TALHHLRIVG CETTHFSVKL FEDLAALRRL ELDQISTAST SFEMTEDVLM PLARLEKFSL TRSRNIELPQ RLLCSLPHLQ VLNISSNELP SLRREESCVA QQLLIVDLSR NRLTNIEQFL RGIPAIRQIS VAYNSIAELD LSLATPFLQQ LDAEANRIVD LTSLPGTVVH VNLAGNALKR VPDAVAELAS LVALNVSRNE IEAGNSSVFS SPELEMLDAS YNKLDSLPVE WLQKCEKRIA HLHLEHNSIE QLTGGVLANA TNLQTLDLSS NQLRVFRDEV LPENSKIGNL RLSNNSLELL EPSSLSGLKL ESLDLSHNKL TEVPAAIGKV EQLKKVDLSH NRIAKVYQYV LNKIKQLHTV DLSNNQLQSI GPYIFSDSSE LHSLDVSNNE ISLLFKDAFA RCPKLRKISM KMNKIKSLDE GLTEASGLRR LDVSHNEILV LKWSALPENL EILNADNNDI NLLTAASMSP STANLKSVSL SNNGITIMNA DQIPNSLESL DVSNNRLAKL GKTALAAKSQ LRRLNLKGNL LTVVATESMK VVEAVHPLKV EISENPLICD CQMGWMIGGA KPKVLIQDSE TASCSHAVDG HQIQIQSLSK KDLLCPYKSV CEPECICCQY GNCDCKSVCP ANCRCFRDDQ FNINIVRCHG NSSMVPKREF VVSELPVSAT EIILSGVTLP QLRTHSFIGR LRLQRLHING TGLRSIQPKA FHTLPALKTL DLSDNSLISL SGEEFLKCGE VSQLFLNGNR FSTLSRGIFE KLPNLKYLTL HNNSLEDIPQ VLHSTALSKI SLSSNPLRCD CSGGSQQHLH HRRDPKAHPF WEHNAAEWFS LHRHLVVDFP KVECWENVTK AFLTNDTTVL SAYPPNMGND VFVMPIEEFL RDYNSTICVP FSSGFFGQDP QNSDIQH |
-Macromolecule #2: Latrophilin 1/LAT-1
| Macromolecule | Name: Latrophilin 1/LAT-1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: AVPSNKPTTD ESGTISHTIC DGEAAELSCP AGKVISIVLG NYGRFSVAVC LPDNDIVPSN INCQNHKTKS ILEKKCNGDS MCYFTVDKKT FTEDPCPNTP KYLEVKYNCV VPATTTTTTT TTSTTTTDSS LIVDEEEEAQ KDALNSDVIK PVKKKEDVFC SATNRRGVNW ...String: AVPSNKPTTD ESGTISHTIC DGEAAELSCP AGKVISIVLG NYGRFSVAVC LPDNDIVPSN INCQNHKTKS ILEKKCNGDS MCYFTVDKKT FTEDPCPNTP KYLEVKYNCV VPATTTTTTT TTSTTTTDSS LIVDEEEEAQ KDALNSDVIK PVKKKEDVFC SATNRRGVNW QNTKSGTTSS APCPEGSSGK QLWACTEEGQ WLTEFPNSAG CESNWISSRN SVLSGVISSE DVSGLPEFLR NLGSETRRPM VGGDLPKVLH LLEKTVNVIA EESWAYQHLP LSNKGAVEVM NYMLRNQEIW GSWDVTKRKE FASRFILAAE KAMVASAKGM MTSAESNVIV QPAITVEISH KIKMSSQPTD YILFPSAALW NGQNVDNVNI PRDAILKINK DETQVFFSSF DNLGAQMTPS DVTVAIAGTD QTEVRKRRVV SRIVGASLIE NGKERRVENL TQPVRITFYH KESSVRHLSN PTCVWWNHHE LKWKPSGCKL SYHNKTMTSC DCTHLTHFAV LMDVRGHDLN EIDQTLLTHH HHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.28 mg/mL |
|---|---|
| Buffer | pH: 8.5 / Details: 10 mM Tris, pH 8.5, 150 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 3333 / Average exposure time: 4.2 sec. / Average electron dose: 60.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)