+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MBP-tagged fusion fatty acid synthase from S. cerevisiae | |||||||||
 Map data Map data | 6xMBP fused FAS | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fatty acid synthase / TRANSFERASE | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Mazhab-Jafari MT / Khalili Samani E | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Direct structural analysis of a single acyl carrier protein domain in fatty acid synthase from the fungus Saccharomyces cerevisiae. Authors: Elnaz Khalili Samani / Amy C Chen / Jennifer W Lou / David L Dai / Alexander F A Keszei / Guihong Tan / Charles Boone / Martin Grininger / Mohammad T Mazhab-Jafari /   Abstract: Acyl carrier protein (ACP) is the work horse of polyketide (PKS) and fatty acid synthases (FAS) and acts as a substrate shuttling domain in these mega enzymes. In fungi, FAS forms a 2.6 MDa symmetric ...Acyl carrier protein (ACP) is the work horse of polyketide (PKS) and fatty acid synthases (FAS) and acts as a substrate shuttling domain in these mega enzymes. In fungi, FAS forms a 2.6 MDa symmetric assembly with six identical copies of FAS1 and FAS2 polypeptides. However, ACP spatial distribution is not restricted by symmetry owing to the long and flexible loops that tether the shuttling domain to its corresponding FAS2 polypeptide. This symmetry breaking has hampered experimental investigation of substrate shuttling route in fungal FAS. Here, we develop a protein engineering and expression method to isolate asymmetric fungal FAS proteins containing odd numbers of ACP domains. Electron cryomicroscopy (cryoEM) observation of the engineered complex reveals a non-uniform distribution of the substrate shuttling domain relative to its corresponding FAS2 polypeptide at 2.9 Å resolution. This work lays the methodological foundation for experimental study of ACP shuttling route in fungi. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40783.map.gz emd_40783.map.gz | 122.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40783-v30.xml emd-40783-v30.xml emd-40783.xml emd-40783.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40783.png emd_40783.png | 79.3 KB | ||
| Filedesc metadata |  emd-40783.cif.gz emd-40783.cif.gz | 7.6 KB | ||
| Others |  emd_40783_half_map_1.map.gz emd_40783_half_map_1.map.gz emd_40783_half_map_2.map.gz emd_40783_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40783 http://ftp.pdbj.org/pub/emdb/structures/EMD-40783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40783 | HTTPS FTP |
-Validation report
| Summary document |  emd_40783_validation.pdf.gz emd_40783_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40783_full_validation.pdf.gz emd_40783_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_40783_validation.xml.gz emd_40783_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_40783_validation.cif.gz emd_40783_validation.cif.gz | 19.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40783 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40783 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40783.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40783.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 6xMBP fused FAS | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_40783_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
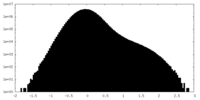
| Projections & Slices |
| ||||||||||||
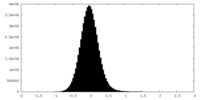
| Density Histograms |
-Half map: Half map B
| File | emd_40783_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
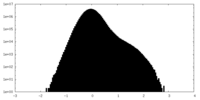
| Projections & Slices |
| ||||||||||||
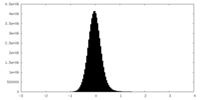
| Density Histograms |
- Sample components
Sample components
-Entire : MBP-tagged Fatty acid synthase
| Entire | Name: MBP-tagged Fatty acid synthase |
|---|---|
| Components |
|
-Supramolecule #1: MBP-tagged Fatty acid synthase
| Supramolecule | Name: MBP-tagged Fatty acid synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.7 MDa |
-Macromolecule #1: Fatty acid synthase
| Macromolecule | Name: Fatty acid synthase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MDAYSTRPLT LSHGSLEHVL LVPTASFFIA SQLQEQFNKI LPEPTEGFAA DDEPTTPAEL VGKFLGYVSS LVEPSKVGQF DQVLNLCLTE FENCYLEGND IHALAAKLLQ ENDTTLVKTK ELIKNYITAR IMAKRPFDKK SNSALFRAVG EGNAQLVAIF GGQGNTDDYF ...String: MDAYSTRPLT LSHGSLEHVL LVPTASFFIA SQLQEQFNKI LPEPTEGFAA DDEPTTPAEL VGKFLGYVSS LVEPSKVGQF DQVLNLCLTE FENCYLEGND IHALAAKLLQ ENDTTLVKTK ELIKNYITAR IMAKRPFDKK SNSALFRAVG EGNAQLVAIF GGQGNTDDYF DELRDLYQTY HVLVGDLIKF SAETLSELIR TTLDAEKVFT QGLNILEWLE NPSNTPDKDY LLSIPISCPL IGVIQLAHYV VTAKLLGFTP GELRSYLKGA TGHSQGLVTA VAIAETDSWE SFFVSVRKAI TVLFFIGVRC YEAYPNTSLP PSILEDSLEN NEGVPSPMLS ISNLTQEQVQ DYVNKTNSHL PAGKQVEISL VNGAKNLVVS GPPQSLYGLN LTLRKAKAPS GLDQSRIPFS ERKLKFSNRF LPVASPFHSH LLVPASDLIN KDLVKNNVSF NAKDIQIPVY DTFDGSDLRV LSGSISERIV DCIIRLPVKW ETTTQFKATH ILDFGPGGAS GLGVLTHRNK DGTGVRVIVA GTLDINPDDD YGFKQEIFDV TSNGLKKNPN WLEEYHPKLI KNKSGKIFVE TKFSKLIGRP PLLVPGMTPC TVSPDFVAAT TNAGYTIELA GGGYFSAAGM TAAIDSVVSQ IEKGSTFGIN LIYVNPFMLQ WGIPLIKELR SKGYPIQFLT IGAGVPSLEV ASEYIETLGL KYLGLKPGSI DAISQVINIA KAHPNFPIAL QWTGGRGGGH HSFEDAHTPM LQMYSKIRRH PNIMLIFGSG FGSADDTYPY LTGEWSTKFD YPPMPFDGFL FGSRVMIAKE VKTSPDAKKC IAACTGVPDD KWEQTYKKPT GGIVTVRSEM GEPIHKIATR GVMLWKEFDE TIFNLPKNKL VPTLEAKRDY IISRLNADFQ KPWFATVNGQ ARDLATMTYE EVAKRLVELM FIRSTNSWFD VTWRTFTGDF LRRVEERFTK SKTLSLIQSY SLLDKPDEAI EKVFNAYPAA REQFLNAQDI DHFLSMCQNP MQKPVPFVPV LDRRFEIFFK KDSLWQSEHL EAVVDQDVQR TCILHGPVAA QFTKVIDEPI KSIMDGIHDG HIKKLLHQYY GDDESKIPAV EYFGGESPVD VQSQVDSSSV SEDSAVFKAT SSTDEESWFK ALAGSEINWR HASFLCSFIT QDKMFVSNPI RKVFKPSQGM VVEISNGNTS SKTVVTLSEP VQGGSGSGSG SMKIEEGKLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE HPDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAKGKSALM FNLQEPYFTW PLIAADGGYA FKYENGKYDI KDVGVDNAGA KAGLTFLVDL IKNKHMNADT DYSIAEAAFN KGETAMTING PWAWSNIDTS KVNYGVTVLP TFKGQPSKPF VGVLSAGINA ASPNKELAKE FLENYLLTDE GLEAVNKDKP LGAVALKSYE EELAKDPRIA ATMENAQKGE IMPNIPQMSA FWYAVRTAVI NAASGRQTVD EALKDAQTGS GSGSGSELKP TVILKLLKEN IIQMEMIENR TMDGKPVSLP LLYNFNPDNG FAPISEVMED RNQRIKEMYW KLWIDEPFNL DFDPRDVIKG KDFEITAKEV YDFTHAVGNN CEDFVSRPDR TMLAPMDFAI VVGWRAIIKA IFPNTVDGDL LKLVHLSNGY KMIPGAKPLQ VGDVVSTTAV IESVVNQPTG KIVDVVGTLS RNGKPVMEVT SSFFYRGNYT DFENTFQKTV EPVYQMHIKT SKDIAVLRSK EWFQLDDEDF DLLNKTLTFE TETEVTFKNA NIFSSVKCFG PIKVELPTKE TVEIGIVDYE AGASHGNPVV DFLKRNGSTL EQKVNLENPI PIAVLDSYTP STNEPYARVS GDLNPIHVSR HFASYANLPG TITHGMFSSA SVRALIENWA ADSVSSRVRG YTCQFVDMVL PNTALKTSIQ HVGMINGRKL IKFETRNEDD VVVLTGEAEI EQPVTTFVFT GQGSQEQGMG MDLYKTSKAA QDVWNRADNH FKDTYGFSIL DIVINNPVNL TIHFGGEKGK RIRENYSAMI FETIVDGKLK TEKIFKEINE HSTSYTFRSE KGLLSATQFT QPALTLMEKA AFEDLKSKGL IPADATFAGH SLGEYAALAS LADVMSIESL VEVVFYRGMT MQVAVPRDEL GRSNYGMIAI NPGRVAASFS QEALQYVVER VGKRTGWLVE IVNYNVENQQ YVAAGDLRAL DTVTNVLNFI KLQKIDIIEL QKSLSLEEVE GHLFEIIDEA SKKSAVKPRP LKLERGFACI PLVGISVPFH STYLMNGVKP FKSFLKKNII KENVKVARLA GKYIPNLTAK PFQVTKEYFQ DVYDLTGSEP IKEIIDNWEK DEWGSPEVEQ ELAHILLTEL LAYQFASPVR WIETQDVFLK DFNTERVVEI GPSPTLAGMA QRTLKNKYES YDAALSLHRE ILCYSKDAKE IYYTPDPSEL AAKEEPAKEE APAPTPAASA PAPAAAAPAP VAAAAPAAAA AEIADEPVKA SLLLHVLVAH KLKKSLDSIP MSKTIKDLVG GKSTVQNEIL GDLGKEFGTT PEKPEETPLE ELAETFQDTF SGALGKQSSS LLSRLISSKM PGGFTITVAR KYLQTRWGLP SGRQDGVLLV ALSNEPAARL GSEADAKAFL DSMAQKYASI VGVDLSSAAS ASGAAGAGAA AGAAMIDAGA LEEITKDHKV LARQQLQVLA RYLKMDLDNG ERKFLKEKDT VAELQAQLDY LNAELGEFFV NGVATSFSRK KARTFDSSWN WAKQSLLSLY FEIIHGVLKN VDREVVSEAI NIMNRSNDAL IKFMEYHISN TDETKGENYQ LVKTLGEQLI ENCKQVLDVD PVYKDVAKPT GPKTAIDKNG NITYSEEPRE KVRKLSQYVQ EMALGGPITK ESQPTIEEDL TRVYKAISAQ ADKQDISSST RVEFEKLYSD LMKFLESSKE IDPSQTTQLA GMDVEDALDK DSTKEVASLP NKSTISKTVS STIPRETIPF LHLRKKTPAG DWKYDRQLSS LFLDGLEKAA FNGVTFKDKY VLITGAGKGS IGAEVLQGLL QGGAKVVVTT SRFSKQVTDY YQSIYAKYGA KGSTLIVVPF NQGSKQDVEA LIEFIYDTEK NGGLGWDLDA IIPFAAIPEQ GIELEHIDSK SEFAHRIMLT NILRMMGCVK KQKSARGIET RPAQVILPMS PNHGTFGGDG MYSESKLSLE TLFNRWHSES WANQLTVCGA IIGWTRGTGL MSANNIIAEG IEKMGVRTFS QKEMAFNLLG LLTPEVVELC QKSPVMADLN GGLQFVPELK EFTAKLRKEL VETSEVRKAV SIETALEHKV VNGNSADAAY AQVEIQPRAN IQLDFPELKP YKQVKQIAPA ELEGLLDLER VIVVTGFAEV GPWGSARTRW EMEAFGEFSL EGCVEMAWIM GFISYHNGNL KGRPYTGWVD SKTKEPVDDK DVKAKYETSI LEHSGIRLIE PELFNGYNPE KKEMIQEVIV EEDLEPFEAS KETAEQFKHQ HGDKVDIFEI PETGEYSVKL LKGATLYIPK ALRFDRLVAG QIPTGWNAKT YGISDDIISQ VDPITLFVLV SVVEAFIASG ITDPYEMYKY VHVSEVGNCS GSGMGGVSAL RGMFKDRFKD EPVQNDILQE SFINTMSAWV NMLLISSSGP IKTPVGACAT SVESVDIGVE TILSGKARIC IVGGYDDFQE EGSFEFGNMK ATSNTLEEFE HGRTPAEMSR PATTTRNGFM EAQGAGIQII MQADLALKMG VPIYGIVAMA ATATDKIGRS VPAPGKGILT TAREHHSSVK YASPNLNMKY RKRQLVTREA QIKDWVENEL EALKLEAEEI PSEDQNEFLL ERTREIHNEA ESQLRAAQQQ WGNDFYKRDP RIAPLRGALA TYGLTIDDLG VASFHGTSTK ANDKNESATI NEMMKHLGRS EGNPVIGVFQ KFLTGHPKGA AGAWMMNGAL QILNSGIIPG NRNADNVDKI LEQFEYVLYP SKTLKTDGVR AVSITSFGFG QKGGQAIVVH PDYLYGAITE DRYNEYVAKV SAREKSAYKF FHNGMIYNKL FVSKEHAPYT DELEEDVYLD PLARVSKDKK SGSLTFNSKN IQSKDSYINA NTIETAKMIE NMTKEKVSNG GVGVDVELIT SINVENDTFI ERNFTPQEIE YCSAQPSVQS SFAGTWSAKE AVFKSLGVKS LGGGAALKDI EIVRVNKNAP AVELHGNAKK AAEEAGVTDV KVSISHDDLQ AVAVAVSTKK HHHHHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 3974 / Average electron dose: 54.76 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.21 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 4.0.2) / Number images used: 71932 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4) |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)