+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Isobutyryl-CoA mutase fused in the presence of GMPPCP | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | supramolecular complex / b12-binding / G-protein chaperone / mutase / ISOMERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationisobutyryl-CoA mutase / pivalyl-CoA mutase activity / isobutyryl-CoA mutase activity / methylmalonyl-CoA mutase activity / acyl-CoA metabolic process / cobalamin binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / GTPase activity / GTP binding / magnesium ion binding Similarity search - Function | ||||||||||||
| Biological species |  Cupriavidus metallidurans CH34 (bacteria) Cupriavidus metallidurans CH34 (bacteria) | ||||||||||||
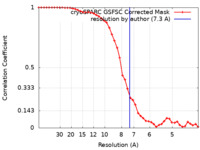
| Method | single particle reconstruction / cryo EM / Resolution: 7.3 Å | ||||||||||||
 Authors Authors | Vaccaro FA / Drennan CL | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: Structural insight into G-protein chaperone-mediated maturation of a bacterial adenosylcobalamin-dependent mutase. Authors: Francesca A Vaccaro / Daphne A Faber / Gisele A Andree / David A Born / Gyunghoon Kang / Dallas R Fonseca / Marco Jost / Catherine L Drennan /  Abstract: G-protein metallochaperones are essential for the proper maturation of numerous metalloenzymes. The G-protein chaperone MMAA in humans (MeaB in bacteria) uses GTP hydrolysis to facilitate the ...G-protein metallochaperones are essential for the proper maturation of numerous metalloenzymes. The G-protein chaperone MMAA in humans (MeaB in bacteria) uses GTP hydrolysis to facilitate the delivery of adenosylcobalamin (AdoCbl) to AdoCbl-dependent methylmalonyl-CoA mutase, an essential metabolic enzyme. This G-protein chaperone also facilitates the removal of damaged cobalamin (Cbl) for repair. Although most chaperones are standalone proteins, isobutyryl-CoA mutase fused (IcmF) has a G-protein domain covalently attached to its target mutase. We previously showed that dimeric MeaB undergoes a 180° rotation to reach a state capable of GTP hydrolysis (an active G-protein state), in which so-called switch III residues of one protomer contact the G-nucleotide of the other protomer. However, it was unclear whether other G-protein chaperones also adopted this conformation. Here, we show that the G-protein domain in a fused system forms a similar active conformation, requiring IcmF oligomerization. IcmF oligomerizes both upon Cbl damage and in the presence of the nonhydrolyzable GTP analog, guanosine-5'-[(β,γ)-methyleno]triphosphate, forming supramolecular complexes observable by mass photometry and EM. Cryo-EM structural analysis reveals that the second protomer of the G-protein intermolecular dimer props open the mutase active site using residues of switch III as a wedge, allowing for AdoCbl insertion or damaged Cbl removal. With the series of structural snapshots now available, we now describe here the molecular basis of G-protein-assisted AdoCbl-dependent mutase maturation, explaining how GTP binding prepares a mutase for cofactor delivery and how GTP hydrolysis allows the mutase to capture the cofactor. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40758.map.gz emd_40758.map.gz | 427.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40758-v30.xml emd-40758-v30.xml emd-40758.xml emd-40758.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40758_fsc.xml emd_40758_fsc.xml | 4.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40758.png emd_40758.png | 48.7 KB | ||
| Others |  emd_40758_half_map_1.map.gz emd_40758_half_map_1.map.gz emd_40758_half_map_2.map.gz emd_40758_half_map_2.map.gz | 7.3 MB 7.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40758 http://ftp.pdbj.org/pub/emdb/structures/EMD-40758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40758 | HTTPS FTP |
-Validation report
| Summary document |  emd_40758_validation.pdf.gz emd_40758_validation.pdf.gz | 651.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40758_full_validation.pdf.gz emd_40758_full_validation.pdf.gz | 650.9 KB | Display | |
| Data in XML |  emd_40758_validation.xml.gz emd_40758_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_40758_validation.cif.gz emd_40758_validation.cif.gz | 14 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40758 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40758 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40758 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40758 | HTTPS FTP |
-Related structure data
| Related structure data |  8staMC  8sslC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40758.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40758.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.0143 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40758_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40758_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Supramolecular complexes of isobutyryl-CoA mutase fused
| Entire | Name: Supramolecular complexes of isobutyryl-CoA mutase fused |
|---|---|
| Components |
|
-Supramolecule #1: Supramolecular complexes of isobutyryl-CoA mutase fused
| Supramolecule | Name: Supramolecular complexes of isobutyryl-CoA mutase fused type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Cupriavidus metallidurans CH34 (bacteria) Cupriavidus metallidurans CH34 (bacteria) |
-Macromolecule #1: Isobutyryl-CoA mutase fused
| Macromolecule | Name: Isobutyryl-CoA mutase fused / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: methylmalonyl-CoA mutase |
|---|---|
| Source (natural) | Organism:  Cupriavidus metallidurans CH34 (bacteria) Cupriavidus metallidurans CH34 (bacteria) |
| Molecular weight | Theoretical: 122.927586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MTDLSDVSRT AAAKPPAVPG RGPANKVRFV TAASLFDGHD ASINIMRRIL QSQGCEVIHL GHNRSVQEV VTAALQEDVQ GIAISSYQGG HVEYFKYMID LLREHGGEHI QVFGGGGGVI VPDEIRELQA YGVARIYSPE D GQRMGLAG ...String: MGSSHHHHHH SSGLVPRGSH MTDLSDVSRT AAAKPPAVPG RGPANKVRFV TAASLFDGHD ASINIMRRIL QSQGCEVIHL GHNRSVQEV VTAALQEDVQ GIAISSYQGG HVEYFKYMID LLREHGGEHI QVFGGGGGVI VPDEIRELQA YGVARIYSPE D GQRMGLAG MITDMAQRCD IDLTRYAPTT LDTVVAGDRR ALAQLITALE NGKADPELVS ALHAQAKAAA VPVLGITGTG GA GKSSLTD ELIRRFRLDQ DDALSIAVIS IDPSRRKSGG ALLGDRIRMN AINHPNIFMR SLATREAGSE ISQALPDVIA ACK AARFDL VIVETSGIGQ GDAAIVPHVD LSLYVMTPEF GAASQLEKID MLDFADFVAI NKFDRKGAQD AWRDVAKQVQ RNRE QWHSR AEDMPVYGTQ ASRFNDDGVT MLYQGLVGAL GARGMSLKPG TLPNLEGRIS TGQNVIVPPA RSRYLAELAD TVRAY HRRV VAQSKLARER QQLRAAHDML QGAGHESAAL ETLASERDVS LGAVERKLLA MWPQMQQAYS GDEYVVKIRD KEIRTG LIS TTLSGTKIRK VVLPRFEDEG EILKWLMREN VPGSFPYTAG VFAFKREGED PTRMFAGEGD AFRTNRRFKL VSEGMEA KR LSTAFDSVTL YGEDPHERPD IYGKVGNSGV SIATLEDMKV LYDGFDLTNP STSVSMTING PAPTILAMFM NTAIDQQI D RFRADNGRDP TADEEAKIRA WVLQNVRGTV QADILKEDQG QNTCIFSTEF SLKVMGDIQE YFVHHQVRNF YSVSISGYH IAEAGANPIS QLAFTLANGF TYVEAYLARG MHIDDFAPNL SFFFSNGMDP EYSVLGRVAR RIWAVTMRDK YGANDRSQKL KYHIQTSGR SLHAQEIDFN DIRTTLQALI AIYDNCNSLH TNAYDEAITT PTAESVRRAL AIQLIINREW GVAKCENPNQ G SFLIEELT DLVEEAVLQE FERIAERGGV LGAMETGYQR GKIQEESLYY EQLKHDGTLP IIGVNTFRNP NGDPTPQTLE LA RSSEDEK QSQLHRLTEF HGAHQADAEA MLARLRQAVI DNRNVFAVLM DAVRVCSLGQ ITHALFEVGG QYRRNM UniProtKB: Fused isobutyryl-CoA mutase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES pH 8, 50 mM NaCl, 500 mM butyryl-CoA, 500 mM GMPPCP, 500 mM MgCl2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Pressure: 1.0 kPa Details: After glow discharge, the grid was coated with graphene oxide prior to use. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | In the presence of the non-hydrolyzable GMPPCP, filamentous supramolecular complexes are formed |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 602 / Average exposure time: 3.99 sec. / Average electron dose: 42.81 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: B / Chain - Residue range: 22-1093 / Chain - Source name: PDB / Chain - Initial model type: experimental model Details: The initial model consisted of two different fragments: resi 22-442 and resi 443-1093 |
|---|---|
| Details | Initial local fitting was done using ChimeraX and then Phenix real space refine was used for rigid body refinement of the defined fragments. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8sta: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)