[English] 日本語
 Yorodumi
Yorodumi- EMDB-40659: Cryo-EM structure of human CST bound to POT1(ESDL)/TPP1 in the ab... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human CST bound to POT1(ESDL)/TPP1 in the absence of telomeric ssDNA | |||||||||
 Map data Map data | Sharpened full map of human CST bound to POT1(ESDL)/TPP1 in the absence of telomeric ssDNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | telomere / shelterin / cst / complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of DNA strand elongation / positive regulation of telomeric D-loop disassembly / G-rich single-stranded DNA binding / telomere assembly / CST complex / segmentation / urogenital system development / 8-hydroxy-2'-deoxyguanosine DNA binding / telomeric D-loop binding / protection from non-homologous end joining at telomere ...positive regulation of DNA strand elongation / positive regulation of telomeric D-loop disassembly / G-rich single-stranded DNA binding / telomere assembly / CST complex / segmentation / urogenital system development / 8-hydroxy-2'-deoxyguanosine DNA binding / telomeric D-loop binding / protection from non-homologous end joining at telomere / regulation of double-strand break repair via nonhomologous end joining / telomerase inhibitor activity / DEAD/H-box RNA helicase binding / establishment of protein localization to telomere / regulation of establishment of protein localization to telomere / telomeric D-loop disassembly / shelterin complex / telomere maintenance via telomere lengthening / Telomere C-strand synthesis initiation / regulation of telomere maintenance via telomerase / Telomere C-strand (Lagging Strand) Synthesis / single-stranded telomeric DNA binding / nuclear telomere cap complex / G-rich strand telomeric DNA binding / telomere capping / intermediate filament cytoskeleton / Processive synthesis on the C-strand of the telomere / Polymerase switching on the C-strand of the telomere / Removal of the Flap Intermediate from the C-strand / telomerase holoenzyme complex / embryonic limb morphogenesis / bone marrow development / protein localization to chromosome, telomeric region / telomeric DNA binding / hematopoietic stem cell proliferation / negative regulation of telomere maintenance via telomerase / positive regulation of telomere maintenance / Telomere Extension By Telomerase / replicative senescence / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / telomere maintenance via telomerase / Packaging Of Telomere Ends / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / spleen development / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / regulation of G2/M transition of mitotic cell cycle / DNA polymerase binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / positive regulation of telomere maintenance via telomerase / Inhibition of DNA recombination at telomere / Meiotic synapsis / telomere maintenance / bioluminescence / thymus development / positive regulation of DNA replication / skeletal system development / generation of precursor metabolites and energy / intracellular protein transport / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / multicellular organism growth / DNA Damage/Telomere Stress Induced Senescence / host cell cytoplasmic vesicle membrane / fibrillar center / positive regulation of fibroblast proliferation / single-stranded DNA binding / nucleoside-triphosphate phosphatase / outer membrane-bounded periplasmic space / channel activity / monoatomic ion transmembrane transport / chromosome, telomeric region / DNA replication / RNA helicase activity / nuclear body / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / intracellular membrane-bounded organelle / DNA-templated transcription / DNA damage response / protein-containing complex binding / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
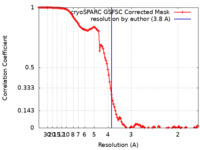
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Cai SW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: POT1 recruits and regulates CST-Polα/Primase at human telomeres. Authors: Sarah W Cai / Hiroyuki Takai / Thomas Walz / Titia de Lange /  Abstract: Telomere maintenance requires extension of the G-rich telomeric repeat strand by telomerase and fill-in synthesis of the C-rich strand by Polα/Primase. Telomeric Polα/Primase is bound to Ctc1-Stn1- ...Telomere maintenance requires extension of the G-rich telomeric repeat strand by telomerase and fill-in synthesis of the C-rich strand by Polα/Primase. Telomeric Polα/Primase is bound to Ctc1-Stn1-Ten1 (CST), a single-stranded DNA-binding complex. Like mutations in telomerase, mutations affecting CST-Polα/Primase result in pathological telomere shortening and cause a telomere biology disorder, Coats plus (CP). We determined cryogenic electron microscopy structures of human CST bound to the shelterin heterodimer POT1/TPP1 that reveal how CST is recruited to telomeres by POT1. Phosphorylation of POT1 is required for CST recruitment, and the complex is formed through conserved interactions involving several residues mutated in CP. Our structural and biochemical data suggest that phosphorylated POT1 holds CST-Polα/Primase in an inactive auto-inhibited state until telomerase has extended the telomere ends. We propose that dephosphorylation of POT1 releases CST-Polα/Primase into an active state that completes telomere replication through fill-in synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40659.map.gz emd_40659.map.gz | 157.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40659-v30.xml emd-40659-v30.xml emd-40659.xml emd-40659.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40659_fsc.xml emd_40659_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40659.png emd_40659.png | 125.8 KB | ||
| Masks |  emd_40659_msk_1.map emd_40659_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40659.cif.gz emd-40659.cif.gz | 8.2 KB | ||
| Others |  emd_40659_half_map_1.map.gz emd_40659_half_map_1.map.gz emd_40659_half_map_2.map.gz emd_40659_half_map_2.map.gz | 154.6 MB 154.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40659 http://ftp.pdbj.org/pub/emdb/structures/EMD-40659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40659 | HTTPS FTP |
-Related structure data
| Related structure data |  8sojMC  8sokC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40659.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40659.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened full map of human CST bound to POT1(ESDL)/TPP1 in the absence of telomeric ssDNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
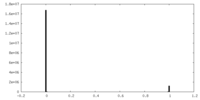
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40659_msk_1.map emd_40659_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
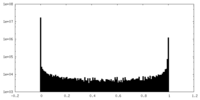
| Density Histograms |
-Half map: Half map A of human CST bound to...
| File | emd_40659_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of human CST bound to POT1(ESDL)/TPP1 in the absence of telomeric ssDNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
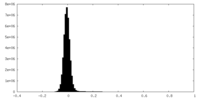
| Density Histograms |
-Half map: Half map B of human CST bound to...
| File | emd_40659_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of human CST bound to POT1(ESDL)/TPP1 in the absence of telomeric ssDNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
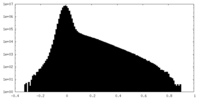
| Density Histograms |
- Sample components
Sample components
-Entire : Human CST-POT1(ESDL)/TPP1 complex
| Entire | Name: Human CST-POT1(ESDL)/TPP1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Human CST-POT1(ESDL)/TPP1 complex
| Supramolecule | Name: Human CST-POT1(ESDL)/TPP1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: CST-POT1(ESDL)/TPP1 complex in the absence of DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 380 KDa |
-Macromolecule #1: CST complex subunit CTC1
| Macromolecule | Name: CST complex subunit CTC1 / type: protein_or_peptide / ID: 1 / Details: His6-MBP-3C tagged human Ctc1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 178.300609 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSSHHHHHH SSGTKIEEGK LVIWINGDKG YNGLAEVGKK FEKDTGIKVT VEHPDKLEEK FPQVAATGDG PDIIFWAHDR FGGYAQSGL LAEITPDKAF QDKLYPFTWD AVRYNGKLIA YPIAVEALSL IYNKDLLPNP PKTWEEIPAL DKELKAKGKS A LMFNLQEP ...String: MGSSHHHHHH SSGTKIEEGK LVIWINGDKG YNGLAEVGKK FEKDTGIKVT VEHPDKLEEK FPQVAATGDG PDIIFWAHDR FGGYAQSGL LAEITPDKAF QDKLYPFTWD AVRYNGKLIA YPIAVEALSL IYNKDLLPNP PKTWEEIPAL DKELKAKGKS A LMFNLQEP YFTWPLIAAD GGYAFKYENG KYDIKDVGVD NAGAKAGLTF LVDLIKNKHM NADTDYSIAE AAFNKGETAM TI NGPWAWS NIDTSKVNYG VTVLPTFKGQ PSKPFVGVLS AGINAASPNK ELAKEFLENY LLTDEGLEAV NKDKPLGAVA LKS YEEELA KDPRIAATME NAQKGEIMPN IPQMSAFWYA VRTAVINAAS GRQTVDEALK DAQTKLVEKY LEVLFQGPGS MAAG RAQVP SSEQAWLEDA QVFIQKTLCP AVKEPNVQLT PLVIDCVKTV WLSQGRNQGS TLPLSYSFVS VQDLKTHQRL PCCSH LSWS SSAYQAWAQE AGPNGNPLPR EQLLLLGTLT DLSADLEQEC RNGSLYVRDN TGVLSCELID LDLSWLGHLF LFPRWS YLP PARWNSSGEG HLELWDAPVP VFPLTISPGP VTPIPVLYPE SASCLLRLRN KLRGVQRNLA GSLVRLSALV KSKQKAY FI LSLGRSHPAV THVSIIVQVP AQLVWHRALR PGTAYVLTEL RVSKIRGQRQ HVWMTSQSSR LLLLKPECVQ ELELELEG P LLEADPKPLP MPSNSEDKKD PESLVRYSRL LSYSGAVTGV LNEPAGLYEL DGQLGLCLAY QQFRGLRRVM RPGVCLQLQ DVHLLQSVGG GTRRPVLAPC LRGAVLLQSF SRQKPGAHSS RQAYGASLYE QLVWERQLGL PLYLWATKAL EELACKLCPH VLRHHQFLQ HSSPGSPSLG LQLLAPTLDL LAPPGSPVRN AHNEILEEPH HCPLQKYTRL QTPSSFPTLA TLKEEGQRKA W ASFDPKAL LPLPEASYLP SCQLNRRLAW SWLCLLPSAF CPAQVLLGVL VASSHKGCLQ LRDQSGSLPC LLLAKHSQPL SD PRLIGCL VRAERFQLIV ERDVRSSFPS WKELSMPGFI QKQQARVYVQ FFLADALILP VPRPCLHSAT PSTPQTDPTG PEG PHLGQS RLFLLCHKEA LMKRNFCVPP GASPEVPKPA LSFYVLGSWL GGTQRKEGTG WGLPEPQGND DNDQKVHLIF FGSS VRWFE FLHPGQVYRL IAPGPATPML FEKDGSSCIS RRPLELAGCA SCLTVQDNWT LELESSQDIQ DVLDANKSLP ESSLT DLLS DNFTDSLVSF SAEILSRTLC EPLVASLWMK LGNTGAMRRC VKLTVALETA ECEFPPHLDV YIEDPHLPPS LGLLPG ARV HFSQLEKRVS RSHNVYCCFR SSTYVQVLSF PPETTISIPL PHIYLAELLQ GGQSPFQATA SCHIVSVFSL QLFWVCA YC TSICRQGKCT RLGSTCPTQT AISQAIIRLL VEDGTAEAVV TCRNHHVAAA LGLCPREWAS LLDFVQVPGR VVLQFAGP G AQLESSARVD EPMTMFLWTL CTSPSVLRPI VLSFELERKP SKIVPLEPPR LQRFQCGELP FLTHVNPRLR LSCLSIRES EYSSSLGILA SSC UniProtKB: Maltose/maltodextrin-binding periplasmic protein, CST complex subunit CTC1 |
-Macromolecule #2: CST complex subunit STN1
| Macromolecule | Name: CST complex subunit STN1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 115.627211 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWSHPQFEKG GGSGGGSGGS AWSHPQFEKG SVSKGEELFT GVVPILVELD GDVNGHKFSV SGEGEGDATY GKLTLKFICT TGKLPVPWP TLVTTLTYGV QCFSRYPDHM KQHDFFKSAM PEGYVQERTI FFKDDGNYKT RAEVKFEGDT LVNRIELKGI D FKEDGNIL ...String: MWSHPQFEKG GGSGGGSGGS AWSHPQFEKG SVSKGEELFT GVVPILVELD GDVNGHKFSV SGEGEGDATY GKLTLKFICT TGKLPVPWP TLVTTLTYGV QCFSRYPDHM KQHDFFKSAM PEGYVQERTI FFKDDGNYKT RAEVKFEGDT LVNRIELKGI D FKEDGNIL GHKLEYNYNS HNVYIMADKQ KNGIKVNFKI RHNIEDGSVQ LADHYQQNTP IGDGPVLLPD NHYLSTQSAL SK DPNEKRD HMVLLEFVTA AGITLGMDEL YKENLYFQGG SMAGSGRLVL RPWIRELILG SETPSSPRAG QLLEVLQDAE AAV AGPSHA PDTSDVGATL LVSDGTHSVR CLVTREALDT SDWEEKEFGF RGTEGRLLLL QDCGVHVQVA EGGAPAEFYL QVDR FSLLP TEQPRLRVPG CNQDLDVQKK LYDCLEEHLS ESTSSNAGLS LSQLLDEMRE DQEHQGALVC LAESCLTLEG PCTAP PVTH WAASRCKATG EAVYTVPSSM LCISENDQLI LSSLGPCQRT QGPELPPPDP ALQDLSLTLI ASPPSSPSSS GTPALP GHM SSEESGTSIS LLPALSLAAP DPGQRSSSQP SPAICSAPAT LTPRSPHASR TPSSPLQSCT PSLSPRSHVP SPHQALV TR PQKPSLEFKE FVGLPCKNRP PFPRTGATRG AQEGGSGGSM QPGSSRCEEE TPSLLWGLDP VFLAFAKLYI RDILDMKE S RQVPGVFLYN GHPIKQVDVL GTVIGVRERD AFYSYGVDDS TGVINCICWK KLNTESVSAA PSAARELSLT SQLKKLQET IEQKTKIEIG DTIRVRGSIR TYREEREIHA TTYYKVDDPV WNIQIARMLE LPTIYRKVYD QPFHSSALEK EEALSNPGAL DLPSLTSLL SEKAKEFLME NRVQSFYQQE LEMVESLLSL ANQPVIHSAS SDQVNFKKDT TSKAIHSIFK NAIQLLQEKG L VFQKDDGF DNLYYVTRED KDLHRKIHRI IQQDCQKPNH MEKGCHFLHI LACARLSIRP GLSEAVLQQV LELLEDQSDI VS TMEHYYT AF UniProtKB: Genome polyprotein, Adrenocortical dysplasia protein homolog, CST complex subunit STN1 |
-Macromolecule #3: CST complex subunit TEN1
| Macromolecule | Name: CST complex subunit TEN1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.872013 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MMLPKPGTYY LPWEVSAGQV PDGSTLRTFG RLCLYDMIQS RVTLMAQHGS DQHQVLVCTK LVEPFHAQVG SLYIVLGELQ HQQDRGSVV KARVLTCVEG MNLPLLEQAI REQRLYKQER GGSQ UniProtKB: CST complex subunit TEN1 |
-Macromolecule #4: Protection of telomeres protein 1
| Macromolecule | Name: Protection of telomeres protein 1 / type: protein_or_peptide / ID: 4 Details: mPOT1b residues ESDL 323-326 are inserted into the human POT1 sequence between S320 and V321 Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.938219 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSS LVPATNYIYT PLNQLKGGTI VNVYGVVKFF KPPYLSKGTD YCSVVTIVDQ TNVKLTCLLF SGNYEALPII YKNGDIVRF HRLKIQVYKK ETQGITSSGF ASLTFEGTLG APIIPRTSSK YFNFTTEDHK MVEALRVWAS THMSPSWTLL K LCDVQPMQ ...String: MHHHHHHGSS LVPATNYIYT PLNQLKGGTI VNVYGVVKFF KPPYLSKGTD YCSVVTIVDQ TNVKLTCLLF SGNYEALPII YKNGDIVRF HRLKIQVYKK ETQGITSSGF ASLTFEGTLG APIIPRTSSK YFNFTTEDHK MVEALRVWAS THMSPSWTLL K LCDVQPMQ YFDLTCQLLG KAEVDGASFL LKVWDGTRTP FPSWRVLIQD LVLEGDLSHI HRLQNLTIDI LVYDNHVHVA RS LKVGSFL RIYSLHTKLQ SMNSENQTML SLEFHLHGGT SYGRGIRVLP ESNSDVDQLK KDLESANLTA NQHSDVICQS EPD DSFPSS GSESDLVSLY EVERCQQLSA TILTDHQYLE RTPLCAILKQ KAPQQYRIRA KLRSYKPRRL FQSVKLHCPK CHLL QEVPH EGDLDIIFQD GATKTPDVKL QNTSLYDSKI WTTKNQKGRK VAVHFVKNNG ILPLSNECLL LIEGGTLSEI CKLSN KFNS VIPVRSGHED LELLDLSAPF LIQGTIHHYG CKQCSSLRSI QNLNSLVDKT SWIPSSVAEA LGIVPLQYVF VMTFTL DDG TGVLEAYLMD SDKFFQIPAS EVLMDDDLQK SVDMIMDMFC PPGIKIDAYP WLECFIKSYN VTNGTDNQIC YQIFDTT VA EDVI UniProtKB: Protection of telomeres protein 1 |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)